Devinder Kumar
Unsupervised Domain Adaptation in Person re-ID via k-Reciprocal Clustering and Large-Scale Heterogeneous Environment Synthesis
Jan 14, 2020
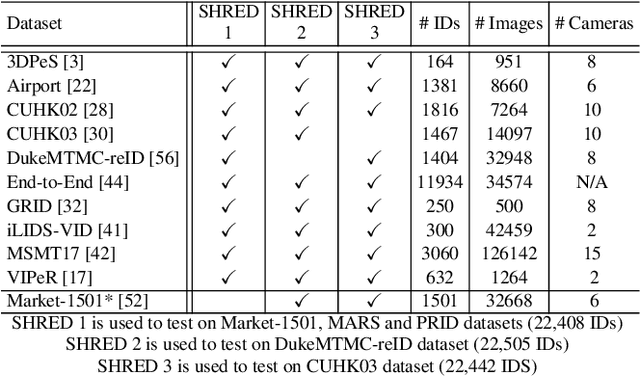
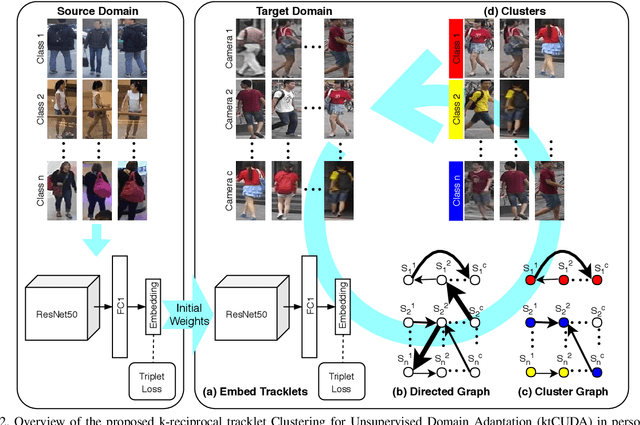
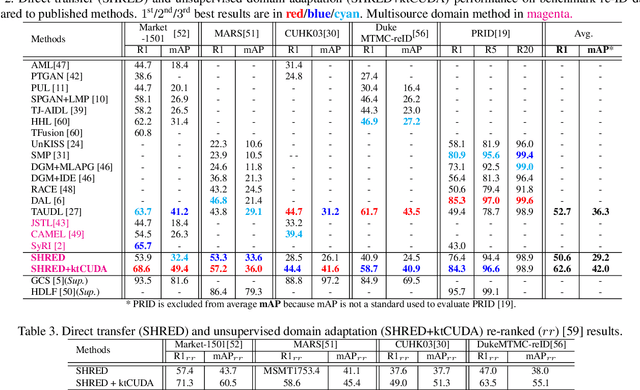
Abstract:An ongoing major challenge in computer vision is the task of person re-identification, where the goal is to match individuals across different, non-overlapping camera views. While recent success has been achieved via supervised learning using deep neural networks, such methods have limited widespread adoption due to the need for large-scale, customized data annotation. As such, there has been a recent focus on unsupervised learning approaches to mitigate the data annotation issue; however, current approaches in literature have limited performance compared to supervised learning approaches as well as limited applicability for adoption in new environments. In this paper, we address the aforementioned challenges faced in person re-identification for real-world, practical scenarios by introducing a novel, unsupervised domain adaptation approach for person re-identification. This is accomplished through the introduction of: i) k-reciprocal tracklet Clustering for Unsupervised Domain Adaptation (ktCUDA) (for pseudo-label generation on target domain), and ii) Synthesized Heterogeneous RE-id Domain (SHRED) composed of large-scale heterogeneous independent source environments (for improving robustness and adaptability to a wide diversity of target environments). Experimental results across four different image and video benchmark datasets show that the proposed ktCUDA and SHRED approach achieves an average improvement of +5.7 mAP in re-identification performance when compared to existing state-of-the-art methods, as well as demonstrate better adaptability to different types of environments.
Fairest of Them All: Establishing a Strong Baseline for Cross-Domain Person ReID
Jul 28, 2019
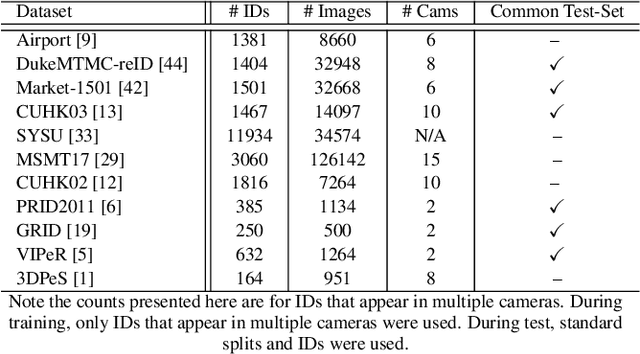

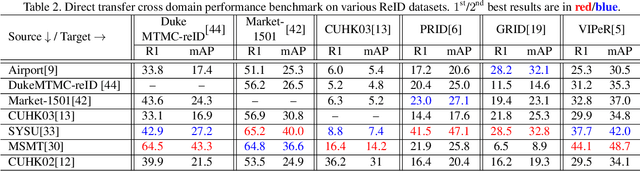
Abstract:Person re-identification (ReID) remains a very difficult challenge in computer vision, and critical for large-scale video surveillance scenarios where an individual could appear in different camera views at different times. There has been recent interest in tackling this challenge using cross-domain approaches, which leverages data from source domains that are different than the target domain. Such approaches are more practical for real-world widespread deployment given that they don't require on-site training (as with unsupervised or domain transfer approaches) or on-site manual annotation and training (as with supervised approaches). In this study, we take a systematic approach to establishing a large baseline source domain and target domain for cross-domain person ReID. We accomplish this by conducting a comprehensive analysis to study the similarities between source domains proposed in literature, and studying the effects of incrementally increasing the size of the source domain. This allows us to establish a balanced source domain and target domain split that promotes variety in both source and target domains. Furthermore, using lessons learned from the state-of-the-art supervised person re-identification methods, we establish a strong baseline method for cross-domain person ReID. Experiments show that a source domain composed of two of the largest person ReID domains (SYSU and MSMT) performs well across six commonly-used target domains. Furthermore, we show that, surprisingly, two of the recent commonly-used domains (PRID and GRID) have too few query images to provide meaningful insights. As such, based on our findings, we propose the following balanced baseline for cross-domain person ReID consisting of: i) a fixed multi-source domain consisting of SYSU, MSMT, Airport and 3DPeS, and ii) a multi-target domain consisting of Market-1501, DukeMTMC-reID, CUHK03, PRID, GRID and VIPeR.
Beyond Explainability: Leveraging Interpretability for Improved Adversarial Learning
Apr 21, 2019



Abstract:In this study, we propose the leveraging of interpretability for tasks beyond purely the purpose of explainability. In particular, this study puts forward a novel strategy for leveraging gradient-based interpretability in the realm of adversarial examples, where we use insights gained to aid adversarial learning. More specifically, we introduce the concept of spatially constrained one-pixel adversarial perturbations, where we guide the learning of such adversarial perturbations towards more susceptible areas identified via gradient-based interpretability. Experimental results using different benchmark datasets show that such a spatially constrained one-pixel adversarial perturbation strategy can noticeably improve the speed of convergence as well as produce successful attacks that were also visually difficult to perceive, thus illustrating an effective use of interpretability methods for tasks outside of the purpose of purely explainability.
SISC: End-to-end Interpretable Discovery Radiomics-Driven Lung Cancer Prediction via Stacked Interpretable Sequencing Cells
Jan 15, 2019



Abstract:Objective: Lung cancer is the leading cause of cancer-related death worldwide. Computer-aided diagnosis (CAD) systems have shown significant promise in recent years for facilitating the effective detection and classification of abnormal lung nodules in computed tomography (CT) scans. While hand-engineered radiomic features have been traditionally used for lung cancer prediction, there have been significant recent successes achieving state-of-the-art results in the area of discovery radiomics. Here, radiomic sequencers comprising of highly discriminative radiomic features are discovered directly from archival medical data. However, the interpretation of predictions made using such radiomic sequencers remains a challenge. Method: A novel end-to-end interpretable discovery radiomics-driven lung cancer prediction pipeline has been designed, build, and tested. The radiomic sequencer being discovered possesses a deep architecture comprised of stacked interpretable sequencing cells (SISC). Results: The SISC architecture is shown to outperform previous approaches while providing more insight in to its decision making process. Conclusion: The SISC radiomic sequencer is able to achieve state-of-the-art results in lung cancer prediction, and also offers prediction interpretability in the form of critical response maps. Significance: The critical response maps are useful for not only validating the predictions of the proposed SISC radiomic sequencer, but also provide improved radiologist-machine collaboration for effective diagnosis.
Understanding Anatomy Classification Through Attentive Response Maps
Feb 07, 2018



Abstract:One of the main challenges for broad adoption of deep learning based models such as convolutional neural networks (CNN), is the lack of understanding of their decisions. In many applications, a simpler, less capable model that can be easily understood is favorable to a black-box model that has superior performance. In this paper, we present an approach for designing CNNs based on visualization of the internal activations of the model. We visualize the model's response through attentive response maps obtained using a fractional stride convolution technique and compare the results with known imaging landmarks from the medical literature. We show that sufficiently deep and capable models can be successfully trained to use the same medical landmarks a human expert would use. Our approach allows for communicating the model decision process well, but also offers insight towards detecting biases.
Discovery Radiomics with CLEAR-DR: Interpretable Computer Aided Diagnosis of Diabetic Retinopathy
Oct 29, 2017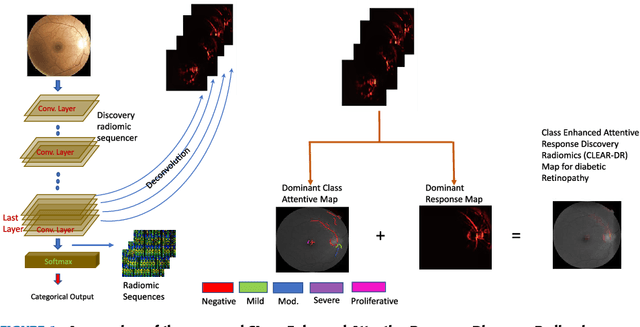
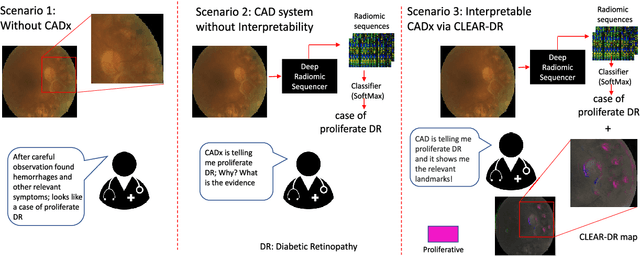
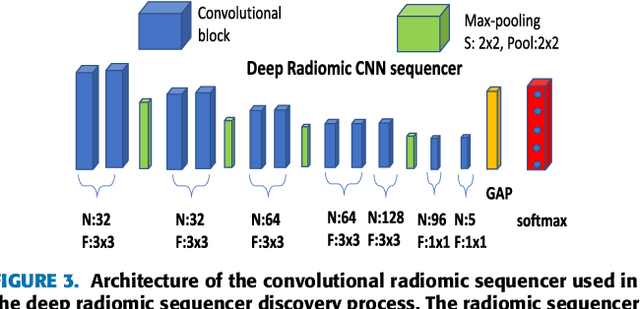
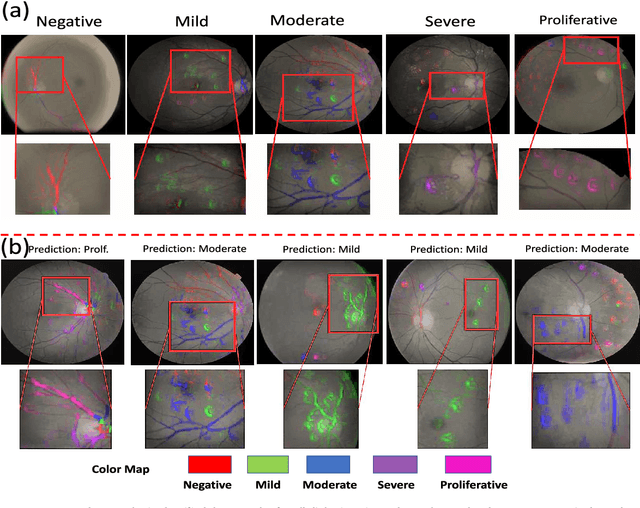
Abstract:Objective: Radiomics-driven Computer Aided Diagnosis (CAD) has shown considerable promise in recent years as a potential tool for improving clinical decision support in medical oncology, particularly those based around the concept of Discovery Radiomics, where radiomic sequencers are discovered through the analysis of medical imaging data. One of the main limitations with current CAD approaches is that it is very difficult to gain insight or rationale as to how decisions are made, thus limiting their utility to clinicians. Methods: In this study, we propose CLEAR-DR, a novel interpretable CAD system based on the notion of CLass-Enhanced Attentive Response Discovery Radiomics for the purpose of clinical decision support for diabetic retinopathy. Results: In addition to disease grading via the discovered deep radiomic sequencer, the CLEAR-DR system also produces a visual interpretation of the decision-making process to provide better insight and understanding into the decision-making process of the system. Conclusion: We demonstrate the effectiveness and utility of the proposed CLEAR-DR system of enhancing the interpretability of diagnostic grading results for the application of diabetic retinopathy grading. Significance: CLEAR-DR can act as a potential powerful tool to address the uninterpretability issue of current CAD systems, thus improving their utility to clinicians.
Opening the Black Box of Financial AI with CLEAR-Trade: A CLass-Enhanced Attentive Response Approach for Explaining and Visualizing Deep Learning-Driven Stock Market Prediction
Sep 05, 2017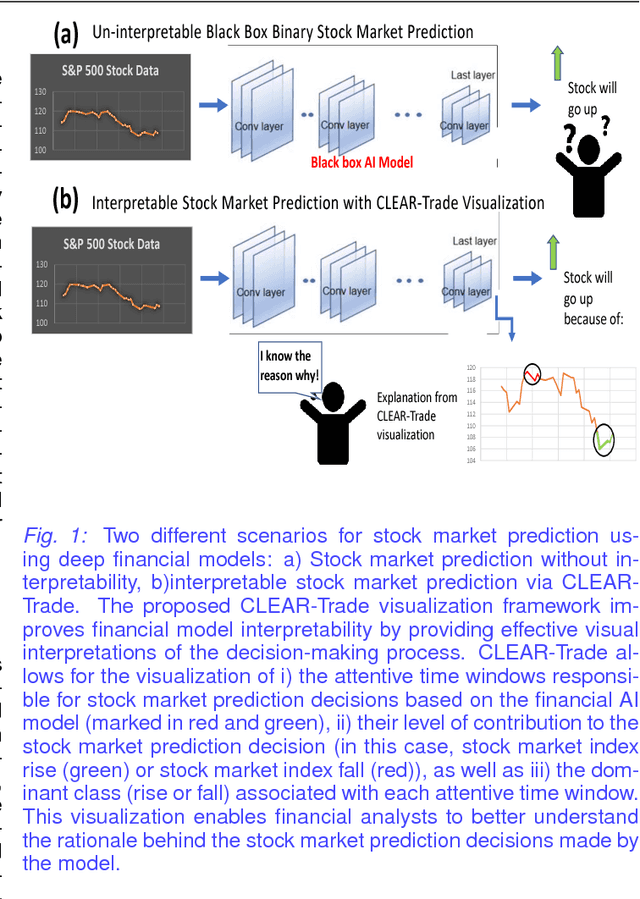
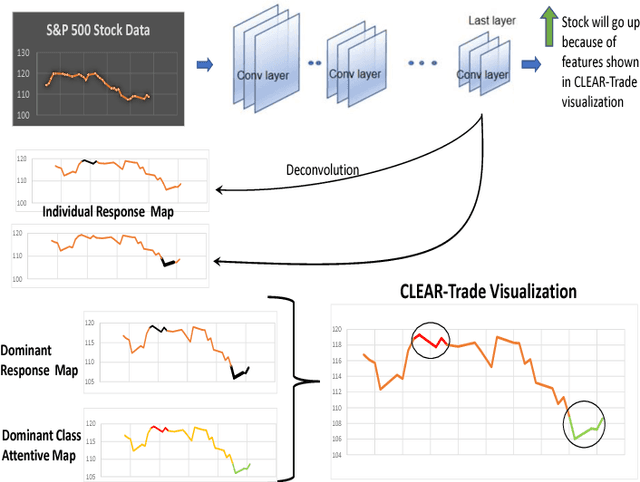
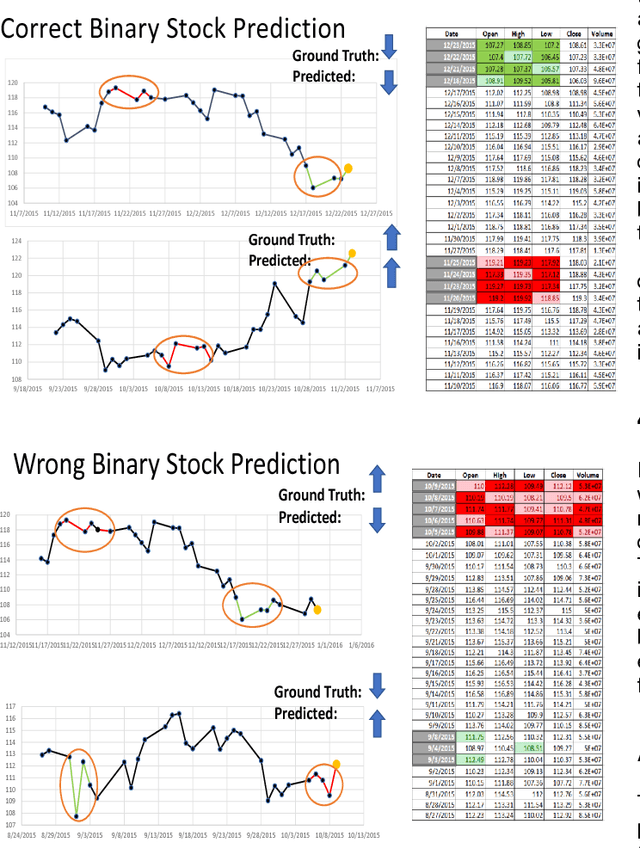
Abstract:Deep learning has been shown to outperform traditional machine learning algorithms across a wide range of problem domains. However, current deep learning algorithms have been criticized as uninterpretable "black-boxes" which cannot explain their decision making processes. This is a major shortcoming that prevents the widespread application of deep learning to domains with regulatory processes such as finance. As such, industries such as finance have to rely on traditional models like decision trees that are much more interpretable but less effective than deep learning for complex problems. In this paper, we propose CLEAR-Trade, a novel financial AI visualization framework for deep learning-driven stock market prediction that mitigates the interpretability issue of deep learning methods. In particular, CLEAR-Trade provides a effective way to visualize and explain decisions made by deep stock market prediction models. We show the efficacy of CLEAR-Trade in enhancing the interpretability of stock market prediction by conducting experiments based on S&P 500 stock index prediction. The results demonstrate that CLEAR-Trade can provide significant insight into the decision-making process of deep learning-driven financial models, particularly for regulatory processes, thus improving their potential uptake in the financial industry.
Explaining the Unexplained: A CLass-Enhanced Attentive Response (CLEAR) Approach to Understanding Deep Neural Networks
May 18, 2017



Abstract:In this work, we propose CLass-Enhanced Attentive Response (CLEAR): an approach to visualize and understand the decisions made by deep neural networks (DNNs) given a specific input. CLEAR facilitates the visualization of attentive regions and levels of interest of DNNs during the decision-making process. It also enables the visualization of the most dominant classes associated with these attentive regions of interest. As such, CLEAR can mitigate some of the shortcomings of heatmap-based methods associated with decision ambiguity, and allows for better insights into the decision-making process of DNNs. Quantitative and qualitative experiments across three different datasets demonstrate the efficacy of CLEAR for gaining a better understanding of the inner workings of DNNs during the decision-making process.
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction
Mar 28, 2017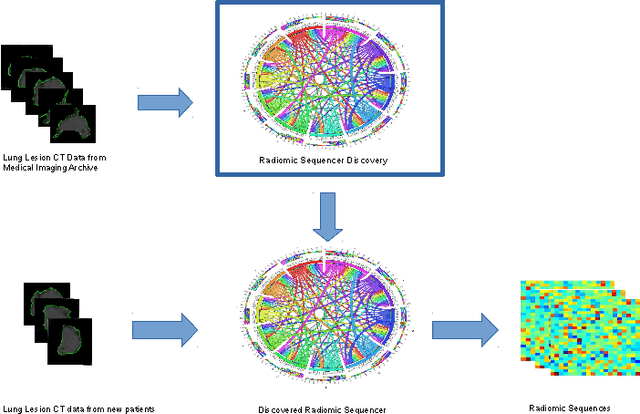
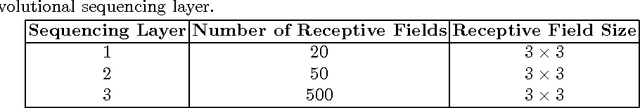
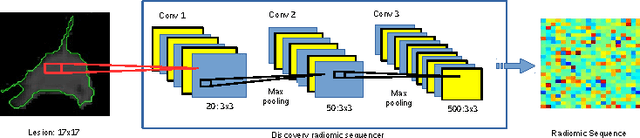
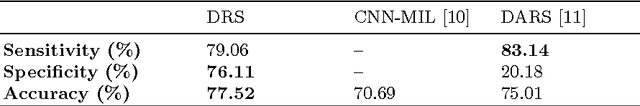
Abstract:Lung cancer is the leading cause for cancer related deaths. As such, there is an urgent need for a streamlined process that can allow radiologists to provide diagnosis with greater efficiency and accuracy. A powerful tool to do this is radiomics: a high-dimension imaging feature set. In this study, we take the idea of radiomics one step further by introducing the concept of discovery radiomics for lung cancer prediction using CT imaging data. In this study, we realize these custom radiomic sequencers as deep convolutional sequencers using a deep convolutional neural network learning architecture. To illustrate the prognostic power and effectiveness of the radiomic sequences produced by the discovered sequencer, we perform cancer prediction between malignant and benign lesions from 97 patients using the pathologically-proven diagnostic data from the LIDC-IDRI dataset. Using the clinically provided pathologically-proven data as ground truth, the proposed framework provided an average accuracy of 77.52% via 10-fold cross-validation with a sensitivity of 79.06% and specificity of 76.11%, surpassing the state-of-the art method.
Discovery Radiomics via StochasticNet Sequencers for Cancer Detection
Nov 11, 2015
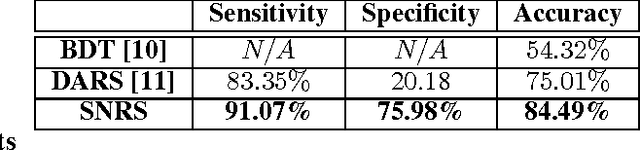
Abstract:Radiomics has proven to be a powerful prognostic tool for cancer detection, and has previously been applied in lung, breast, prostate, and head-and-neck cancer studies with great success. However, these radiomics-driven methods rely on pre-defined, hand-crafted radiomic feature sets that can limit their ability to characterize unique cancer traits. In this study, we introduce a novel discovery radiomics framework where we directly discover custom radiomic features from the wealth of available medical imaging data. In particular, we leverage novel StochasticNet radiomic sequencers for extracting custom radiomic features tailored for characterizing unique cancer tissue phenotype. Using StochasticNet radiomic sequencers discovered using a wealth of lung CT data, we perform binary classification on 42,340 lung lesions obtained from the CT scans of 93 patients in the LIDC-IDRI dataset. Preliminary results show significant improvement over previous state-of-the-art methods, indicating the potential of the proposed discovery radiomics framework for improving cancer screening and diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge