Anuj Srivastava
Graph Network Modeling Techniques for Visualizing Human Mobility Patterns
Apr 04, 2025



Abstract:Human mobility analysis at urban-scale requires models to represent the complex nature of human movements, which in turn are affected by accessibility to nearby points of interest, underlying socioeconomic factors of a place, and local transport choices for people living in a geographic region. In this work, we represent human mobility and the associated flow of movements as a grapyh. Graph-based approaches for mobility analysis are still in their early stages of adoption and are actively being researched. The challenges of graph-based mobility analysis are multifaceted - the lack of sufficiently high-quality data to represent flows at high spatial and teporal resolution whereas, limited computational resources to translate large voluments of mobility data into a network structure, and scaling issues inherent in graph models etc. The current study develops a methodology by embedding graphs into a continuous space, which alleviates issues related to fast graph matching, graph time-series modeling, and visualization of mobility dynamics. Through experiments, we demonstrate how mobility data collected from taxicab trajectories could be transformed into network structures and patterns of mobility flow changes, and can be used for downstream tasks reporting approx 40% decrease in error on average in matched graphs vs unmatched ones.
Investigating Image Manifolds of 3D Objects: Learning, Shape Analysis, and Comparisons
Mar 09, 2025Abstract:Despite high-dimensionality of images, the sets of images of 3D objects have long been hypothesized to form low-dimensional manifolds. What is the nature of such manifolds? How do they differ across objects and object classes? Answering these questions can provide key insights in explaining and advancing success of machine learning algorithms in computer vision. This paper investigates dual tasks -- learning and analyzing shapes of image manifolds -- by revisiting a classical problem of manifold learning but from a novel geometrical perspective. It uses geometry-preserving transformations to map the pose image manifolds, sets of images formed by rotating 3D objects, to low-dimensional latent spaces. The pose manifolds of different objects in latent spaces are found to be nonlinear, smooth manifolds. The paper then compares shapes of these manifolds for different objects using Kendall's shape analysis, modulo rigid motions and global scaling, and clusters objects according to these shape metrics. Interestingly, pose manifolds for objects from the same classes are frequently clustered together. The geometries of image manifolds can be exploited to simplify vision and image processing tasks, to predict performances, and to provide insights into learning methods.
Dynamic Neural Surfaces for Elastic 4D Shape Representation and Analysis
Mar 05, 2025Abstract:We propose a novel framework for the statistical analysis of genus-zero 4D surfaces, i.e., 3D surfaces that deform and evolve over time. This problem is particularly challenging due to the arbitrary parameterizations of these surfaces and their varying deformation speeds, necessitating effective spatiotemporal registration. Traditionally, 4D surfaces are discretized, in space and time, before computing their spatiotemporal registrations, geodesics, and statistics. However, this approach may result in suboptimal solutions and, as we demonstrate in this paper, is not necessary. In contrast, we treat 4D surfaces as continuous functions in both space and time. We introduce Dynamic Spherical Neural Surfaces (D-SNS), an efficient smooth and continuous spatiotemporal representation for genus-0 4D surfaces. We then demonstrate how to perform core 4D shape analysis tasks such as spatiotemporal registration, geodesics computation, and mean 4D shape estimation, directly on these continuous representations without upfront discretization and meshing. By integrating neural representations with classical Riemannian geometry and statistical shape analysis techniques, we provide the building blocks for enabling full functional shape analysis. We demonstrate the efficiency of the framework on 4D human and face datasets. The source code and additional results are available at https://4d-dsns.github.io/DSNS/.
* 22 pages, 23 figures, conference paper
A Shape-Based Functional Index for Objective Assessment of Pediatric Motor Function
Jan 02, 2025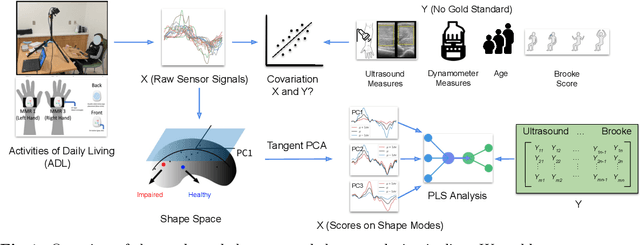
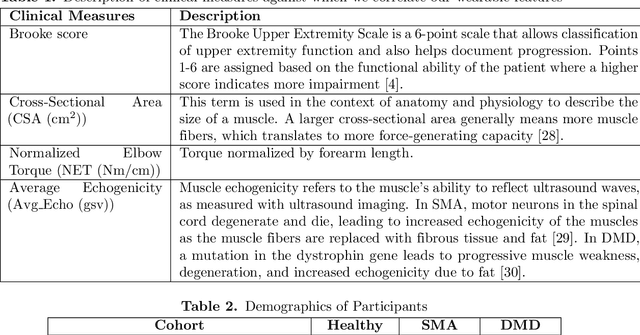
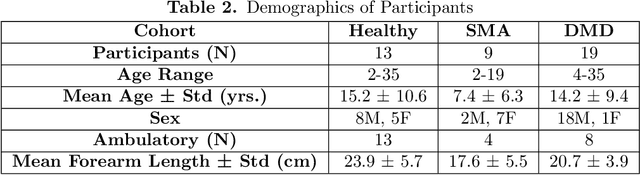
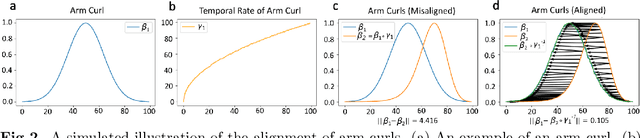
Abstract:Clinical assessments for neuromuscular disorders, such as Spinal Muscular Atrophy (SMA) and Duchenne Muscular Dystrophy (DMD), continue to rely on subjective measures to monitor treatment response and disease progression. We introduce a novel method using wearable sensors to objectively assess motor function during daily activities in 19 patients with DMD, 9 with SMA, and 13 age-matched controls. Pediatric movement data is complex due to confounding factors such as limb length variations in growing children and variability in movement speed. Our approach uses Shape-based Principal Component Analysis to align movement trajectories and identify distinct kinematic patterns, including variations in motion speed and asymmetry. Both DMD and SMA cohorts have individuals with motor function on par with healthy controls. Notably, patients with SMA showed greater activation of the motion asymmetry pattern. We further combined projections on these principal components with partial least squares (PLS) to identify a covariation mode with a canonical correlation of r = 0.78 (95% CI: [0.34, 0.94]) with muscle fat infiltration, the Brooke score (a motor function score), and age-related degenerative changes, proposing a novel motor function index. This data-driven method can be deployed in home settings, enabling better longitudinal tracking of treatment efficacy for children with neuromuscular disorders.
Fused Gromov-Wasserstein Variance Decomposition with Linear Optimal Transport
Nov 15, 2024



Abstract:Wasserstein distances form a family of metrics on spaces of probability measures that have recently seen many applications. However, statistical analysis in these spaces is complex due to the nonlinearity of Wasserstein spaces. One potential solution to this problem is Linear Optimal Transport (LOT). This method allows one to find a Euclidean embedding, called LOT embedding, of measures in some Wasserstein spaces, but some information is lost in this embedding. So, to understand whether statistical analysis relying on LOT embeddings can make valid inferences about original data, it is helpful to quantify how well these embeddings describe that data. To answer this question, we present a decomposition of the Fr\'echet variance of a set of measures in the 2-Wasserstein space, which allows one to compute the percentage of variance explained by LOT embeddings of those measures. We then extend this decomposition to the Fused Gromov-Wasserstein setting. We also present several experiments that explore the relationship between the dimension of the LOT embedding, the percentage of variance explained by the embedding, and the classification accuracy of machine learning classifiers built on the embedded data. We use the MNIST handwritten digits dataset, IMDB-50000 dataset, and Diffusion Tensor MRI images for these experiments. Our results illustrate the effectiveness of low dimensional LOT embeddings in terms of the percentage of variance explained and the classification accuracy of models built on the embedded data.
A Riemannian Approach for Spatiotemporal Analysis and Generation of 4D Tree-shaped Structures
Aug 22, 2024



Abstract:We propose the first comprehensive approach for modeling and analyzing the spatiotemporal shape variability in tree-like 4D objects, i.e., 3D objects whose shapes bend, stretch, and change in their branching structure over time as they deform, grow, and interact with their environment. Our key contribution is the representation of tree-like 3D shapes using Square Root Velocity Function Trees (SRVFT). By solving the spatial registration in the SRVFT space, which is equipped with an L2 metric, 4D tree-shaped structures become time-parameterized trajectories in this space. This reduces the problem of modeling and analyzing 4D tree-like shapes to that of modeling and analyzing elastic trajectories in the SRVFT space, where elasticity refers to time warping. In this paper, we propose a novel mathematical representation of the shape space of such trajectories, a Riemannian metric on that space, and computational tools for fast and accurate spatiotemporal registration and geodesics computation between 4D tree-shaped structures. Leveraging these building blocks, we develop a full framework for modelling the spatiotemporal variability using statistical models and generating novel 4D tree-like structures from a set of exemplars. We demonstrate and validate the proposed framework using real 4D plant data.
Rescuing referral failures during automated diagnosis of domain-shifted medical images
Nov 28, 2023



Abstract:The success of deep learning models deployed in the real world depends critically on their ability to generalize well across diverse data domains. Here, we address a fundamental challenge with selective classification during automated diagnosis with domain-shifted medical images. In this scenario, models must learn to avoid making predictions when label confidence is low, especially when tested with samples far removed from the training set (covariate shift). Such uncertain cases are typically referred to the clinician for further analysis and evaluation. Yet, we show that even state-of-the-art domain generalization approaches fail severely during referral when tested on medical images acquired from a different demographic or using a different technology. We examine two benchmark diagnostic medical imaging datasets exhibiting strong covariate shifts: i) diabetic retinopathy prediction with retinal fundus images and ii) multilabel disease prediction with chest X-ray images. We show that predictive uncertainty estimates do not generalize well under covariate shifts leading to non-monotonic referral curves, and severe drops in performance (up to 50%) at high referral rates (>70%). We evaluate novel combinations of robust generalization and post hoc referral approaches, that rescue these failures and achieve significant performance improvements, typically >10%, over baseline methods. Our study identifies a critical challenge with referral in domain-shifted medical images and finds key applications in reliable, automated disease diagnosis.
Shape-Graph Matching Network (SGM-net): Registration for Statistical Shape Analysis
Aug 14, 2023Abstract:This paper focuses on the statistical analysis of shapes of data objects called shape graphs, a set of nodes connected by articulated curves with arbitrary shapes. A critical need here is a constrained registration of points (nodes to nodes, edges to edges) across objects. This, in turn, requires optimization over the permutation group, made challenging by differences in nodes (in terms of numbers, locations) and edges (in terms of shapes, placements, and sizes) across objects. This paper tackles this registration problem using a novel neural-network architecture and involves an unsupervised loss function developed using the elastic shape metric for curves. This architecture results in (1) state-of-the-art matching performance and (2) an order of magnitude reduction in the computational cost relative to baseline approaches. We demonstrate the effectiveness of the proposed approach using both simulated data and real-world 2D and 3D shape graphs. Code and data will be made publicly available after review to foster research.
Learning Pose Image Manifolds Using Geometry-Preserving GANs and Elasticae
May 17, 2023



Abstract:This paper investigates the challenge of learning image manifolds, specifically pose manifolds, of 3D objects using limited training data. It proposes a DNN approach to manifold learning and for predicting images of objects for novel, continuous 3D rotations. The approach uses two distinct concepts: (1) Geometric Style-GAN (Geom-SGAN), which maps images to low-dimensional latent representations and maintains the (first-order) manifold geometry. That is, it seeks to preserve the pairwise distances between base points and their tangent spaces, and (2) uses Euler's elastica to smoothly interpolate between directed points (points + tangent directions) in the low-dimensional latent space. When mapped back to the larger image space, the resulting interpolations resemble videos of rotating objects. Extensive experiments establish the superiority of this framework in learning paths on rotation manifolds, both visually and quantitatively, relative to state-of-the-art GANs and VAEs.
Data-Driven, Soft Alignment of Functional Data Using Shapes and Landmarks
Apr 10, 2022



Abstract:Alignment or registration of functions is a fundamental problem in statistical analysis of functions and shapes. While there are several approaches available, a more recent approach based on Fisher-Rao metric and square-root velocity functions (SRVFs) has been shown to have good performance. However, this SRVF method has two limitations: (1) it is susceptible to over alignment, i.e., alignment of noise as well as the signal, and (2) in case there is additional information in form of landmarks, the original formulation does not prescribe a way to incorporate that information. In this paper we propose an extension that allows for incorporation of landmark information to seek a compromise between matching curves and landmarks. This results in a soft landmark alignment that pushes landmarks closer, without requiring their exact overlays to finds a compromise between contributions from functions and landmarks. The proposed method is demonstrated to be superior in certain practical scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge