Sathyanarayanan N. Aakur
FSP-DETR: Few-Shot Prototypical Parasitic Ova Detection
Oct 10, 2025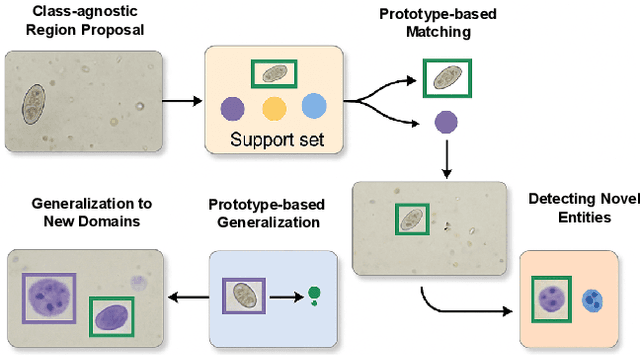
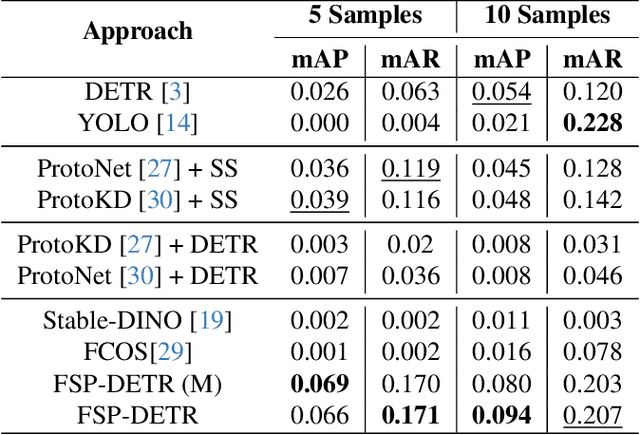
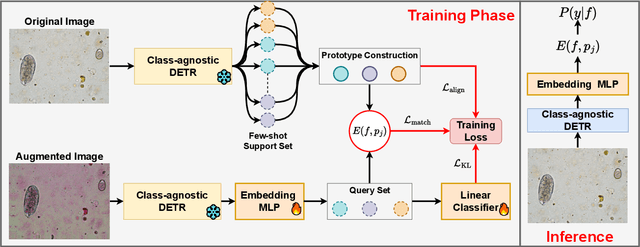
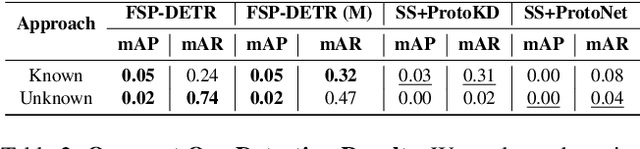
Abstract:Object detection in biomedical settings is fundamentally constrained by the scarcity of labeled data and the frequent emergence of novel or rare categories. We present FSP-DETR, a unified detection framework that enables robust few-shot detection, open-set recognition, and generalization to unseen biomedical tasks within a single model. Built upon a class-agnostic DETR backbone, our approach constructs class prototypes from original support images and learns an embedding space using augmented views and a lightweight transformer decoder. Training jointly optimizes a prototype matching loss, an alignment-based separation loss, and a KL divergence regularization to improve discriminative feature learning and calibration under scarce supervision. Unlike prior work that tackles these tasks in isolation, FSP-DETR enables inference-time flexibility to support unseen class recognition, background rejection, and cross-task adaptation without retraining. We also introduce a new ova species detection benchmark with 20 parasite classes and establish standardized evaluation protocols. Extensive experiments across ova, blood cell, and malaria detection tasks demonstrate that FSP-DETR significantly outperforms prior few-shot and prototype-based detectors, especially in low-shot and open-set scenarios.
STaTS: Structure-Aware Temporal Sequence Summarization via Statistical Window Merging
Oct 10, 2025Abstract:Time series data often contain latent temporal structure, transitions between locally stationary regimes, repeated motifs, and bursts of variability, that are rarely leveraged in standard representation learning pipelines. Existing models typically operate on raw or fixed-window sequences, treating all time steps as equally informative, which leads to inefficiencies, poor robustness, and limited scalability in long or noisy sequences. We propose STaTS, a lightweight, unsupervised framework for Structure-Aware Temporal Summarization that adaptively compresses both univariate and multivariate time series into compact, information-preserving token sequences. STaTS detects change points across multiple temporal resolutions using a BIC-based statistical divergence criterion, then summarizes each segment using simple functions like the mean or generative models such as GMMs. This process achieves up to 30x sequence compression while retaining core temporal dynamics. STaTS operates as a model-agnostic preprocessor and can be integrated with existing unsupervised time series encoders without retraining. Extensive experiments on 150+ datasets, including classification tasks on the UCR-85, UCR-128, and UEA-30 archives, and forecasting on ETTh1 and ETTh2, ETTm1, and Electricity, demonstrate that STaTS enables 85-90\% of the full-model performance while offering dramatic reductions in computational cost. Moreover, STaTS improves robustness under noise and preserves discriminative structure, outperforming uniform and clustering-based compression baselines. These results position STaTS as a principled, general-purpose solution for efficient, structure-aware time series modeling.
Hallucinate, Ground, Repeat: A Framework for Generalized Visual Relationship Detection
Jun 06, 2025Abstract:Understanding relationships between objects is central to visual intelligence, with applications in embodied AI, assistive systems, and scene understanding. Yet, most visual relationship detection (VRD) models rely on a fixed predicate set, limiting their generalization to novel interactions. A key challenge is the inability to visually ground semantically plausible, but unannotated, relationships hypothesized from external knowledge. This work introduces an iterative visual grounding framework that leverages large language models (LLMs) as structured relational priors. Inspired by expectation-maximization (EM), our method alternates between generating candidate scene graphs from detected objects using an LLM (expectation) and training a visual model to align these hypotheses with perceptual evidence (maximization). This process bootstraps relational understanding beyond annotated data and enables generalization to unseen predicates. Additionally, we introduce a new benchmark for open-world VRD on Visual Genome with 21 held-out predicates and evaluate under three settings: seen, unseen, and mixed. Our model outperforms LLM-only, few-shot, and debiased baselines, achieving mean recall (mR@50) of 15.9, 13.1, and 11.7 on predicate classification on these three sets. These results highlight the promise of grounded LLM priors for scalable open-world visual understanding.
A Probabilistic Jump-Diffusion Framework for Open-World Egocentric Activity Recognition
May 28, 2025Abstract:Open-world egocentric activity recognition poses a fundamental challenge due to its unconstrained nature, requiring models to infer unseen activities from an expansive, partially observed search space. We introduce ProbRes, a Probabilistic Residual search framework based on jump-diffusion that efficiently navigates this space by balancing prior-guided exploration with likelihood-driven exploitation. Our approach integrates structured commonsense priors to construct a semantically coherent search space, adaptively refines predictions using Vision-Language Models (VLMs) and employs a stochastic search mechanism to locate high-likelihood activity labels while minimizing exhaustive enumeration efficiently. We systematically evaluate ProbRes across multiple openness levels (L0--L3), demonstrating its adaptability to increasing search space complexity. In addition to achieving state-of-the-art performance on benchmark datasets (GTEA Gaze, GTEA Gaze+, EPIC-Kitchens, and Charades-Ego), we establish a clear taxonomy for open-world recognition, delineating the challenges and methodological advancements necessary for egocentric activity understanding.
ProbRes: Probabilistic Jump Diffusion for Open-World Egocentric Activity Recognition
Apr 04, 2025Abstract:Open-world egocentric activity recognition poses a fundamental challenge due to its unconstrained nature, requiring models to infer unseen activities from an expansive, partially observed search space. We introduce ProbRes, a Probabilistic Residual search framework based on jump-diffusion that efficiently navigates this space by balancing prior-guided exploration with likelihood-driven exploitation. Our approach integrates structured commonsense priors to construct a semantically coherent search space, adaptively refines predictions using Vision-Language Models (VLMs) and employs a stochastic search mechanism to locate high-likelihood activity labels while minimizing exhaustive enumeration efficiently. We systematically evaluate ProbRes across multiple openness levels (L0 - L3), demonstrating its adaptability to increasing search space complexity. In addition to achieving state-of-the-art performance on benchmark datasets (GTEA Gaze, GTEA Gaze+, EPIC-Kitchens, and Charades-Ego), we establish a clear taxonomy for open-world recognition, delineating the challenges and methodological advancements necessary for egocentric activity understanding. Our results highlight the importance of structured search strategies, paving the way for scalable and efficient open-world activity recognition.
Self-supervised Multi-actor Social Activity Understanding in Streaming Videos
Jun 20, 2024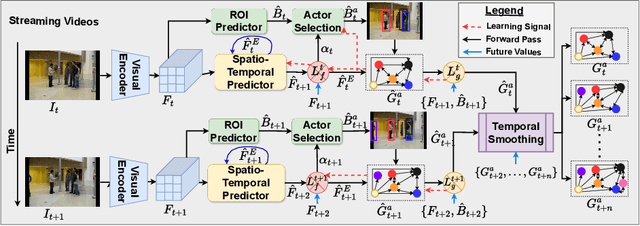
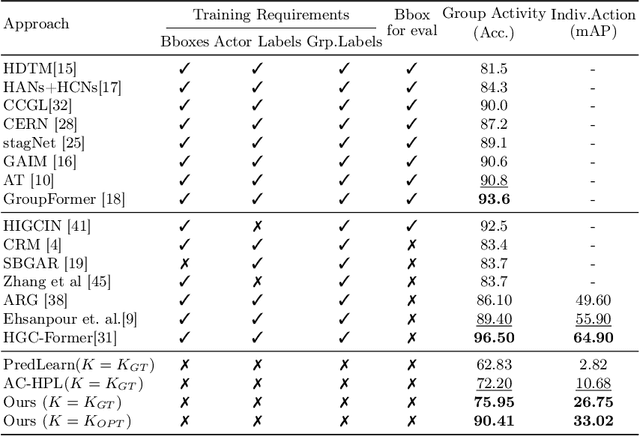
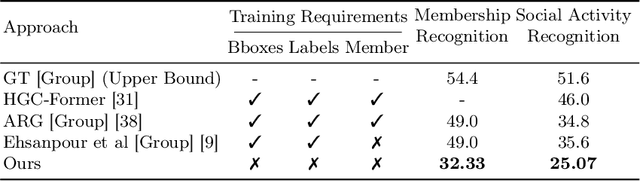

Abstract:This work addresses the problem of Social Activity Recognition (SAR), a critical component in real-world tasks like surveillance and assistive robotics. Unlike traditional event understanding approaches, SAR necessitates modeling individual actors' appearance and motions and contextualizing them within their social interactions. Traditional action localization methods fall short due to their single-actor, single-action assumption. Previous SAR research has relied heavily on densely annotated data, but privacy concerns limit their applicability in real-world settings. In this work, we propose a self-supervised approach based on multi-actor predictive learning for SAR in streaming videos. Using a visual-semantic graph structure, we model social interactions, enabling relational reasoning for robust performance with minimal labeled data. The proposed framework achieves competitive performance on standard group activity recognition benchmarks. Evaluation on three publicly available action localization benchmarks demonstrates its generalizability to arbitrary action localization.
Capturing Temporal Components for Time Series Classification
Jun 20, 2024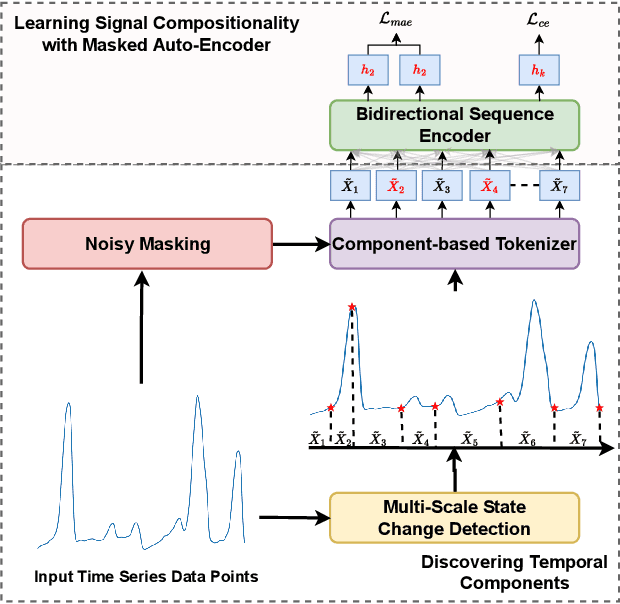
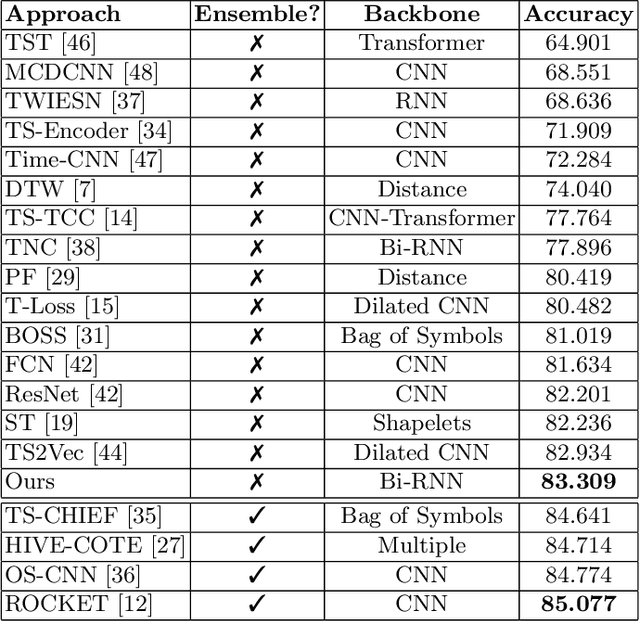
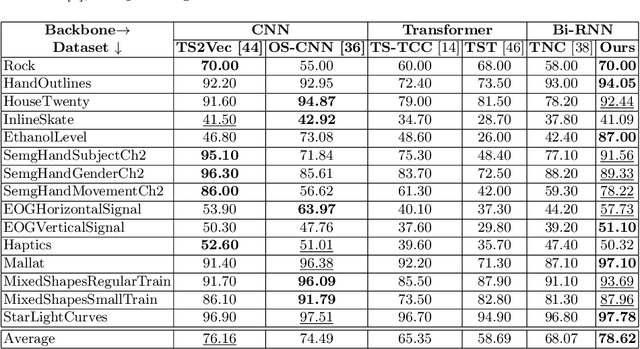
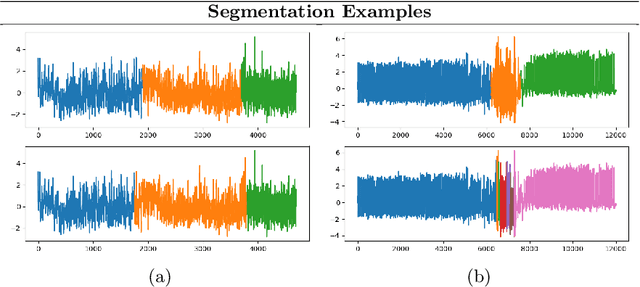
Abstract:Analyzing sequential data is crucial in many domains, particularly due to the abundance of data collected from the Internet of Things paradigm. Time series classification, the task of categorizing sequential data, has gained prominence, with machine learning approaches demonstrating remarkable performance on public benchmark datasets. However, progress has primarily been in designing architectures for learning representations from raw data at fixed (or ideal) time scales, which can fail to generalize to longer sequences. This work introduces a \textit{compositional representation learning} approach trained on statistically coherent components extracted from sequential data. Based on a multi-scale change space, an unsupervised approach is proposed to segment the sequential data into chunks with similar statistical properties. A sequence-based encoder model is trained in a multi-task setting to learn compositional representations from these temporal components for time series classification. We demonstrate its effectiveness through extensive experiments on publicly available time series classification benchmarks. Evaluating the coherence of segmented components shows its competitive performance on the unsupervised segmentation task.
ALGO: Object-Grounded Visual Commonsense Reasoning for Open-World Egocentric Action Recognition
Jun 09, 2024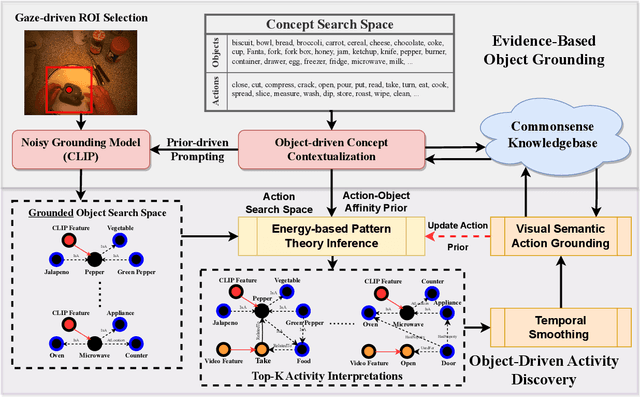
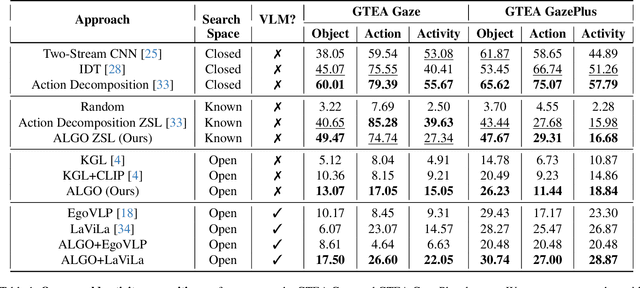
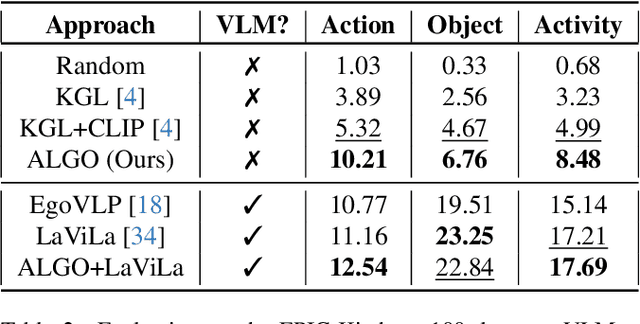
Abstract:Learning to infer labels in an open world, i.e., in an environment where the target "labels" are unknown, is an important characteristic for achieving autonomy. Foundation models pre-trained on enormous amounts of data have shown remarkable generalization skills through prompting, particularly in zero-shot inference. However, their performance is restricted to the correctness of the target label's search space. In an open world, this target search space can be unknown or exceptionally large, which severely restricts the performance of such models. To tackle this challenging problem, we propose a neuro-symbolic framework called ALGO - Action Learning with Grounded Object recognition that uses symbolic knowledge stored in large-scale knowledge bases to infer activities in egocentric videos with limited supervision using two steps. First, we propose a neuro-symbolic prompting approach that uses object-centric vision-language models as a noisy oracle to ground objects in the video through evidence-based reasoning. Second, driven by prior commonsense knowledge, we discover plausible activities through an energy-based symbolic pattern theory framework and learn to ground knowledge-based action (verb) concepts in the video. Extensive experiments on four publicly available datasets (EPIC-Kitchens, GTEA Gaze, GTEA Gaze Plus) demonstrate its performance on open-world activity inference.
ProtoKD: Learning from Extremely Scarce Data for Parasite Ova Recognition
Sep 18, 2023
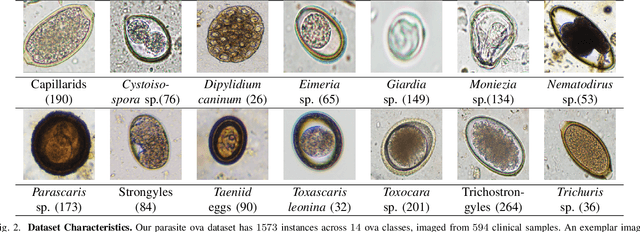
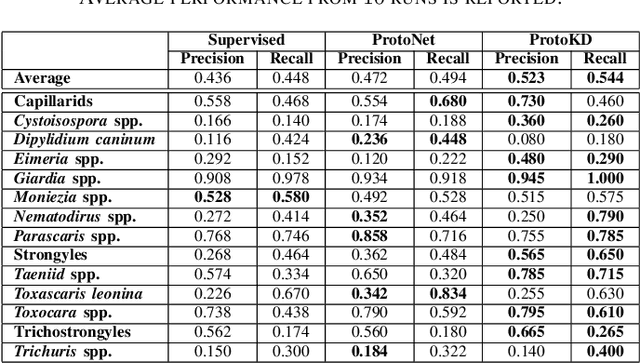
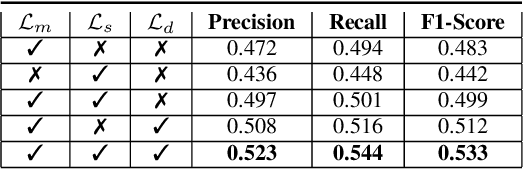
Abstract:Developing reliable computational frameworks for early parasite detection, particularly at the ova (or egg) stage is crucial for advancing healthcare and effectively managing potential public health crises. While deep learning has significantly assisted human workers in various tasks, its application and diagnostics has been constrained by the need for extensive datasets. The ability to learn from an extremely scarce training dataset, i.e., when fewer than 5 examples per class are present, is essential for scaling deep learning models in biomedical applications where large-scale data collection and annotation can be expensive or not possible (in case of novel or unknown infectious agents). In this study, we introduce ProtoKD, one of the first approaches to tackle the problem of multi-class parasitic ova recognition using extremely scarce data. Combining the principles of prototypical networks and self-distillation, we can learn robust representations from only one sample per class. Furthermore, we establish a new benchmark to drive research in this critical direction and validate that the proposed ProtoKD framework achieves state-of-the-art performance. Additionally, we evaluate the framework's generalizability to other downstream tasks by assessing its performance on a large-scale taxonomic profiling task based on metagenomes sequenced from real-world clinical data.
Shape-Graph Matching Network (SGM-net): Registration for Statistical Shape Analysis
Aug 14, 2023Abstract:This paper focuses on the statistical analysis of shapes of data objects called shape graphs, a set of nodes connected by articulated curves with arbitrary shapes. A critical need here is a constrained registration of points (nodes to nodes, edges to edges) across objects. This, in turn, requires optimization over the permutation group, made challenging by differences in nodes (in terms of numbers, locations) and edges (in terms of shapes, placements, and sizes) across objects. This paper tackles this registration problem using a novel neural-network architecture and involves an unsupervised loss function developed using the elastic shape metric for curves. This architecture results in (1) state-of-the-art matching performance and (2) an order of magnitude reduction in the computational cost relative to baseline approaches. We demonstrate the effectiveness of the proposed approach using both simulated data and real-world 2D and 3D shape graphs. Code and data will be made publicly available after review to foster research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge