Priyadharsini Ramamurthy
TEPI: Taxonomy-aware Embedding and Pseudo-Imaging for Scarcely-labeled Zero-shot Genome Classification
Jan 24, 2024Abstract:A species' genetic code or genome encodes valuable evolutionary, biological, and phylogenetic information that aids in species recognition, taxonomic classification, and understanding genetic predispositions like drug resistance and virulence. However, the vast number of potential species poses significant challenges in developing a general-purpose whole genome classification tool. Traditional bioinformatics tools have made notable progress but lack scalability and are computationally expensive. Machine learning-based frameworks show promise but must address the issue of large classification vocabularies with long-tail distributions. In this study, we propose addressing this problem through zero-shot learning using TEPI, Taxonomy-aware Embedding and Pseudo-Imaging. We represent each genome as pseudo-images and map them to a taxonomy-aware embedding space for reasoning and classification. This embedding space captures compositional and phylogenetic relationships of species, enabling predictions in extensive search spaces. We evaluate TEPI using two rigorous zero-shot settings and demonstrate its generalization capabilities qualitatively on curated, large-scale, publicly sourced data.
Scalable Pathogen Detection from Next Generation DNA Sequencing with Deep Learning
Nov 30, 2022Abstract:Next-generation sequencing technologies have enhanced the scope of Internet-of-Things (IoT) to include genomics for personalized medicine through the increased availability of an abundance of genome data collected from heterogeneous sources at a reduced cost. Given the sheer magnitude of the collected data and the significant challenges offered by the presence of highly similar genomic structure across species, there is a need for robust, scalable analysis platforms to extract actionable knowledge such as the presence of potentially zoonotic pathogens. The emergence of zoonotic diseases from novel pathogens, such as the influenza virus in 1918 and SARS-CoV-2 in 2019 that can jump species barriers and lead to pandemic underscores the need for scalable metagenome analysis. In this work, we propose MG2Vec, a deep learning-based solution that uses the transformer network as its backbone, to learn robust features from raw metagenome sequences for downstream biomedical tasks such as targeted and generalized pathogen detection. Extensive experiments on four increasingly challenging, yet realistic diagnostic settings, show that the proposed approach can help detect pathogens from uncurated, real-world clinical samples with minimal human supervision in the form of labels. Further, we demonstrate that the learned representations can generalize to completely unrelated pathogens across diseases and species for large-scale metagenome analysis. We provide a comprehensive evaluation of a novel representation learning framework for metagenome-based disease diagnostics with deep learning and provide a way forward for extracting and using robust vector representations from low-cost next generation sequencing to develop generalizable diagnostic tools.
A Rich Recipe Representation as Plan to Support Expressive Multi Modal Queries on Recipe Content and Preparation Process
Mar 31, 2022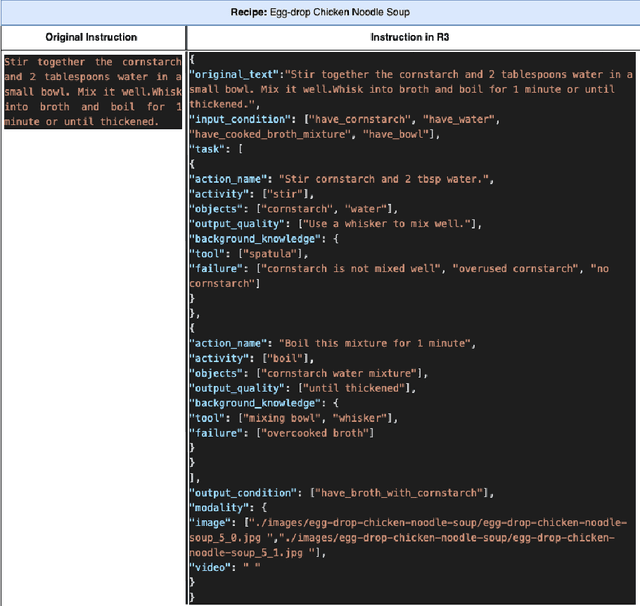
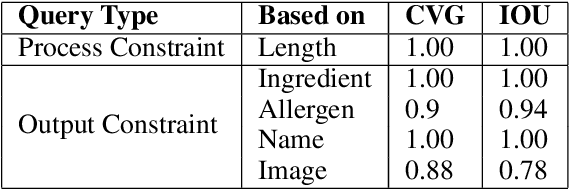
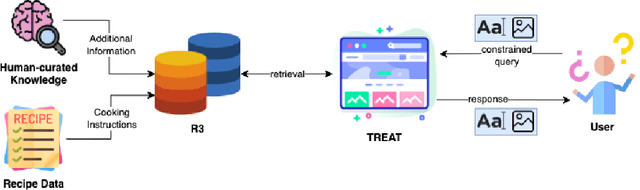
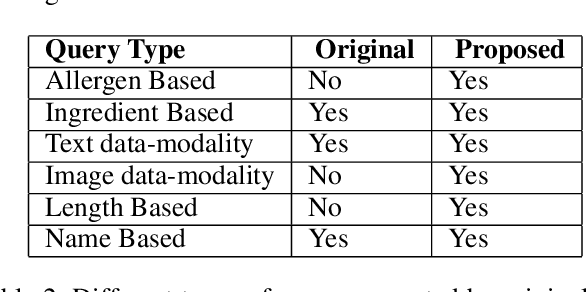
Abstract:Food is not only a basic human necessity but also a key factor driving a society's health and economic well-being. As a result, the cooking domain is a popular use-case to demonstrate decision-support (AI) capabilities in service of benefits like precision health with tools ranging from information retrieval interfaces to task-oriented chatbots. An AI here should understand concepts in the food domain (e.g., recipes, ingredients), be tolerant to failures encountered while cooking (e.g., browning of butter), handle allergy-based substitutions, and work with multiple data modalities (e.g. text and images). However, the recipes today are handled as textual documents which makes it difficult for machines to read, reason and handle ambiguity. This demands a need for better representation of the recipes, overcoming the ambiguity and sparseness that exists in the current textual documents. In this paper, we discuss the construction of a machine-understandable rich recipe representation (R3), in the form of plans, from the recipes available in natural language. R3 is infused with additional knowledge such as information about allergens and images of ingredients, possible failures and tips for each atomic cooking step. To show the benefits of R3, we also present TREAT, a tool for recipe retrieval which uses R3 to perform multi-modal reasoning on the recipe's content (plan objects - ingredients and cooking tools), food preparation process (plan actions and time), and media type (image, text). R3 leads to improved retrieval efficiency and new capabilities that were hither-to not possible in textual representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge