Devarajan Sridharan
Semi-supervised Deep Transfer for Regression without Domain Alignment
Sep 05, 2025Abstract:Deep learning models deployed in real-world applications (e.g., medicine) face challenges because source models do not generalize well to domain-shifted target data. Many successful domain adaptation (DA) approaches require full access to source data. Yet, such requirements are unrealistic in scenarios where source data cannot be shared either because of privacy concerns or because it is too large and incurs prohibitive storage or computational costs. Moreover, resource constraints may limit the availability of labeled targets. We illustrate this challenge in a neuroscience setting where source data are unavailable, labeled target data are meager, and predictions involve continuous-valued outputs. We build upon Contradistinguisher (CUDA), an efficient framework that learns a shared model across the labeled source and unlabeled target samples, without intermediate representation alignment. Yet, CUDA was designed for unsupervised DA, with full access to source data, and for classification tasks. We develop CRAFT -- a Contradistinguisher-based Regularization Approach for Flexible Training -- for source-free (SF), semi-supervised transfer of pretrained models in regression tasks. We showcase the efficacy of CRAFT in two neuroscience settings: gaze prediction with electroencephalography (EEG) data and ``brain age'' prediction with structural MRI data. For both datasets, CRAFT yielded up to 9% improvement in root-mean-squared error (RMSE) over fine-tuned models when labeled training examples were scarce. Moreover, CRAFT leveraged unlabeled target data and outperformed four competing state-of-the-art source-free domain adaptation models by more than 3%. Lastly, we demonstrate the efficacy of CRAFT on two other real-world regression benchmarks. We propose CRAFT as an efficient approach for source-free, semi-supervised deep transfer for regression that is ubiquitous in biology and medicine.
Rescuing referral failures during automated diagnosis of domain-shifted medical images
Nov 28, 2023



Abstract:The success of deep learning models deployed in the real world depends critically on their ability to generalize well across diverse data domains. Here, we address a fundamental challenge with selective classification during automated diagnosis with domain-shifted medical images. In this scenario, models must learn to avoid making predictions when label confidence is low, especially when tested with samples far removed from the training set (covariate shift). Such uncertain cases are typically referred to the clinician for further analysis and evaluation. Yet, we show that even state-of-the-art domain generalization approaches fail severely during referral when tested on medical images acquired from a different demographic or using a different technology. We examine two benchmark diagnostic medical imaging datasets exhibiting strong covariate shifts: i) diabetic retinopathy prediction with retinal fundus images and ii) multilabel disease prediction with chest X-ray images. We show that predictive uncertainty estimates do not generalize well under covariate shifts leading to non-monotonic referral curves, and severe drops in performance (up to 50%) at high referral rates (>70%). We evaluate novel combinations of robust generalization and post hoc referral approaches, that rescue these failures and achieve significant performance improvements, typically >10%, over baseline methods. Our study identifies a critical challenge with referral in domain-shifted medical images and finds key applications in reliable, automated disease diagnosis.
Shaken, and Stirred: Long-Range Dependencies Enable Robust Outlier Detection with PixelCNN++
Aug 29, 2022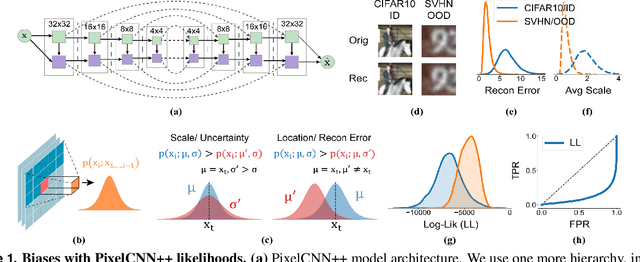
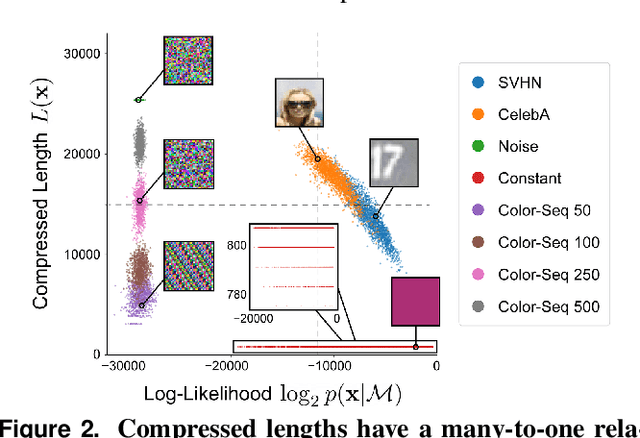
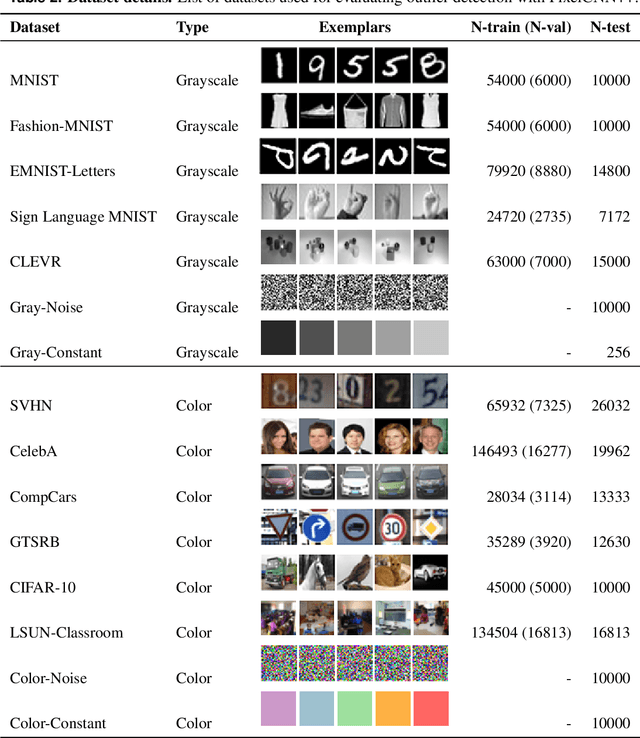
Abstract:Reliable outlier detection is critical for real-world applications of deep learning models. Likelihoods produced by deep generative models, although extensively studied, have been largely dismissed as being impractical for outlier detection. For one, deep generative model likelihoods are readily biased by low-level input statistics. Second, many recent solutions for correcting these biases are computationally expensive or do not generalize well to complex, natural datasets. Here, we explore outlier detection with a state-of-the-art deep autoregressive model: PixelCNN++. We show that biases in PixelCNN++ likelihoods arise primarily from predictions based on local dependencies. We propose two families of bijective transformations that we term "shaking" and "stirring", which ameliorate low-level biases and isolate the contribution of long-range dependencies to the PixelCNN++ likelihood. These transformations are computationally inexpensive and readily applied at evaluation time. We evaluate our approaches extensively with five grayscale and six natural image datasets and show that they achieve or exceed state-of-the-art outlier detection performance. In sum, lightweight remedies suffice to achieve robust outlier detection on images with deep generative models.
Efficient remedies for outlier detection with variational autoencoders
Aug 19, 2021



Abstract:Deep networks often make confident, yet incorrect, predictions when tested with outlier data that is far removed from their training distributions. Likelihoods computed by deep generative models are a candidate metric for outlier detection with unlabeled data. Yet, previous studies have shown that such likelihoods are unreliable and can be easily biased by simple transformations to input data. Here, we examine outlier detection with variational autoencoders (VAEs), among the simplest class of deep generative models. First, we show that a theoretically-grounded correction readily ameliorates a key bias with VAE likelihood estimates. The bias correction is model-free, sample-specific, and accurately computed with the Bernoulli and continuous Bernoulli visible distributions. Second, we show that a well-known preprocessing technique, contrast normalization, extends the effectiveness of bias correction to natural image datasets. Third, we show that the variance of the likelihoods computed over an ensemble of VAEs also enables robust outlier detection. We perform a comprehensive evaluation of our remedies with nine (grayscale and natural) image datasets, and demonstrate significant advantages, in terms of both speed and accuracy, over four other state-of-the-art methods. Our lightweight remedies are biologically inspired and may serve to achieve efficient outlier detection with many types of deep generative models.
Optimizing the Linear Fascicle Evaluation Algorithm for Multi-Core and Many-Core Systems
May 14, 2019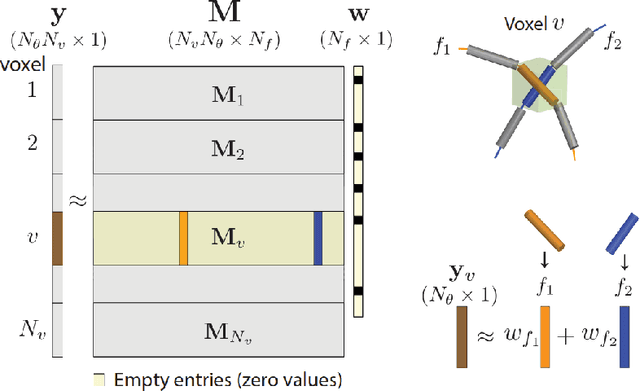
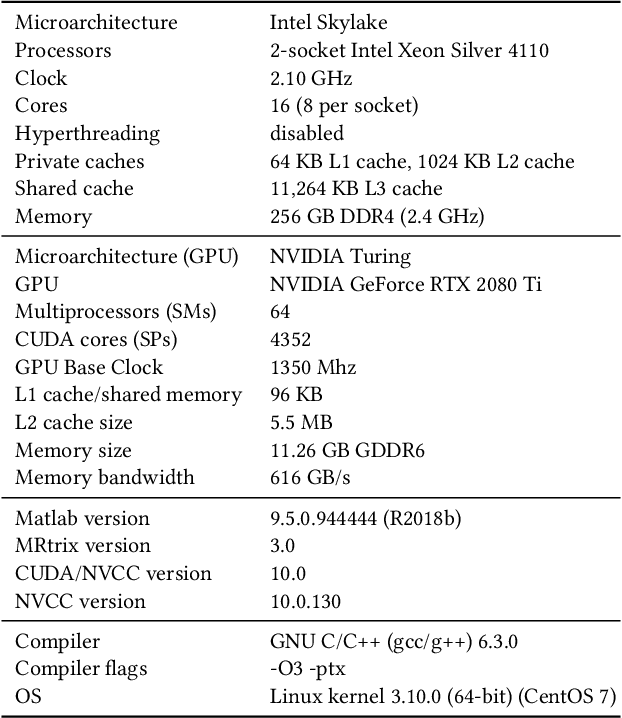
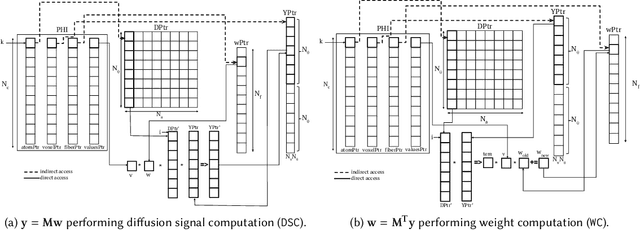
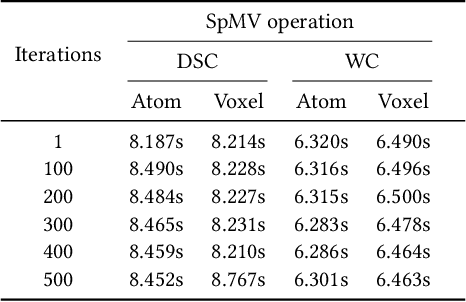
Abstract:Sparse matrix-vector multiplication (SpMV) operations are commonly used in various scientific applications. The performance of the SpMV operation often depends on exploiting regularity patterns in the matrix. Various representations have been proposed to minimize the memory bandwidth bottleneck arising from the irregular memory access pattern involved. Among recent representation techniques, tensor decomposition is a popular one used for very large but sparse matrices. Post sparse-tensor decomposition, the new representation involves indirect accesses, making it challenging to optimize for multi-cores and GPUs. Computational neuroscience algorithms often involve sparse datasets while still performing long-running computations on them. The LiFE application is a popular neuroscience algorithm used for pruning brain connectivity graphs. The datasets employed herein involve the Sparse Tucker Decomposition (STD), a widely used tensor decomposition method. Using this decomposition leads to irregular array references, making it very difficult to optimize for both CPUs and GPUs. Recent codes of the LiFE algorithm show that its SpMV operations are the key bottleneck for performance and scaling. In this work, we first propose target-independent optimizations to optimize these SpMV operations, followed by target-dependent optimizations for CPU and GPU systems. The target-independent techniques include: (1) standard compiler optimizations, (2) data restructuring methods, and (3) methods to partition computations among threads. Then we present the optimizations for CPUs and GPUs to exploit platform-specific speed. Our highly optimized CPU code obtain a speedup of 27.12x over the original sequential CPU code running on 16-core Intel Xeon (Skylake-based) system, and our optimized GPU code achieves a speedup of 5.2x over a reference optimized GPU code version on NVIDIA's GeForce RTX 2080 Ti GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge