Andrew Feng
NeVStereo: A NeRF-Driven NVS-Stereo Architecture for High-Fidelity 3D Tasks
Feb 05, 2026Abstract:In modern dense 3D reconstruction, feed-forward systems (e.g., VGGT, pi3) focus on end-to-end matching and geometry prediction but do not explicitly output the novel view synthesis (NVS). Neural rendering-based approaches offer high-fidelity NVS and detailed geometry from posed images, yet they typically assume fixed camera poses and can be sensitive to pose errors. As a result, it remains non-trivial to obtain a single framework that can offer accurate poses, reliable depth, high-quality rendering, and accurate 3D surfaces from casually captured views. We present NeVStereo, a NeRF-driven NVS-stereo architecture that aims to jointly deliver camera poses, multi-view depth, novel view synthesis, and surface reconstruction from multi-view RGB-only inputs. NeVStereo combines NeRF-based NVS for stereo-friendly renderings, confidence-guided multi-view depth estimation, NeRF-coupled bundle adjustment for pose refinement, and an iterative refinement stage that updates both depth and the radiance field to improve geometric consistency. This design mitigated the common NeRF-based issues such as surface stacking, artifacts, and pose-depth coupling. Across indoor, outdoor, tabletop, and aerial benchmarks, our experiments indicate that NeVStereo achieves consistently strong zero-shot performance, with up to 36% lower depth error, 10.4% improved pose accuracy, 4.5% higher NVS fidelity, and state-of-the-art mesh quality (F1 91.93%, Chamfer 4.35 mm) compared to existing prestigious methods.
LowDiff: Efficient Diffusion Sampling with Low-Resolution Condition
Sep 18, 2025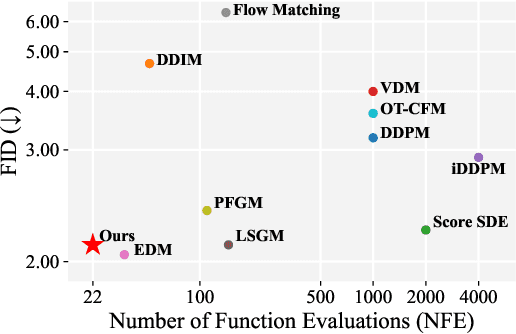
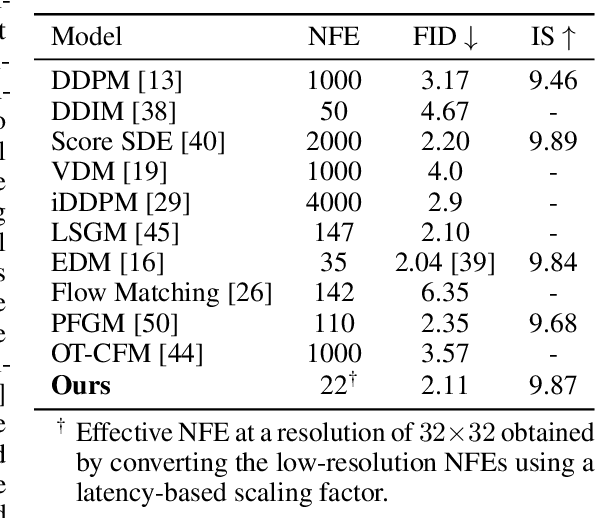
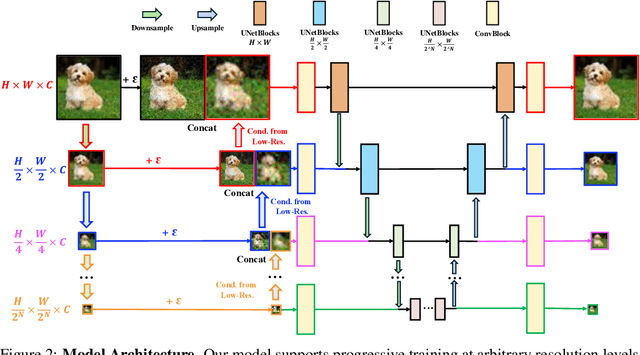
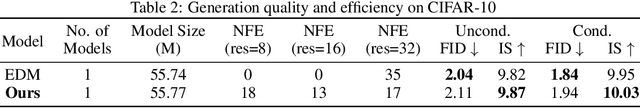
Abstract:Diffusion models have achieved remarkable success in image generation but their practical application is often hindered by the slow sampling speed. Prior efforts of improving efficiency primarily focus on compressing models or reducing the total number of denoising steps, largely neglecting the possibility to leverage multiple input resolutions in the generation process. In this work, we propose LowDiff, a novel and efficient diffusion framework based on a cascaded approach by generating increasingly higher resolution outputs. Besides, LowDiff employs a unified model to progressively refine images from low resolution to the desired resolution. With the proposed architecture design and generation techniques, we achieve comparable or even superior performance with much fewer high-resolution sampling steps. LowDiff is applicable to diffusion models in both pixel space and latent space. Extensive experiments on both conditional and unconditional generation tasks across CIFAR-10, FFHQ and ImageNet demonstrate the effectiveness and generality of our method. Results show over 50% throughput improvement across all datasets and settings while maintaining comparable or better quality. On unconditional CIFAR-10, LowDiff achieves an FID of 2.11 and IS of 9.87, while on conditional CIFAR-10, an FID of 1.94 and IS of 10.03. On FFHQ 64x64, LowDiff achieves an FID of 2.43, and on ImageNet 256x256, LowDiff built on LightningDiT-B/1 produces high-quality samples with a FID of 4.00 and an IS of 195.06, together with substantial efficiency gains.
IDU: Incremental Dynamic Update of Existing 3D Virtual Environments with New Imagery Data
Aug 25, 2025Abstract:For simulation and training purposes, military organizations have made substantial investments in developing high-resolution 3D virtual environments through extensive imaging and 3D scanning. However, the dynamic nature of battlefield conditions-where objects may appear or vanish over time-makes frequent full-scale updates both time-consuming and costly. In response, we introduce the Incremental Dynamic Update (IDU) pipeline, which efficiently updates existing 3D reconstructions, such as 3D Gaussian Splatting (3DGS), with only a small set of newly acquired images. Our approach starts with camera pose estimation to align new images with the existing 3D model, followed by change detection to pinpoint modifications in the scene. A 3D generative AI model is then used to create high-quality 3D assets of the new elements, which are seamlessly integrated into the existing 3D model. The IDU pipeline incorporates human guidance to ensure high accuracy in object identification and placement, with each update focusing on a single new object at a time. Experimental results confirm that our proposed IDU pipeline significantly reduces update time and labor, offering a cost-effective and targeted solution for maintaining up-to-date 3D models in rapidly evolving military scenarios.
Splat Feature Solver
Aug 17, 2025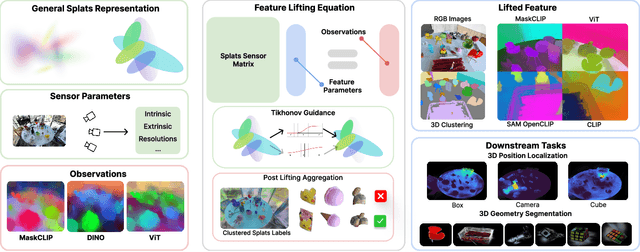
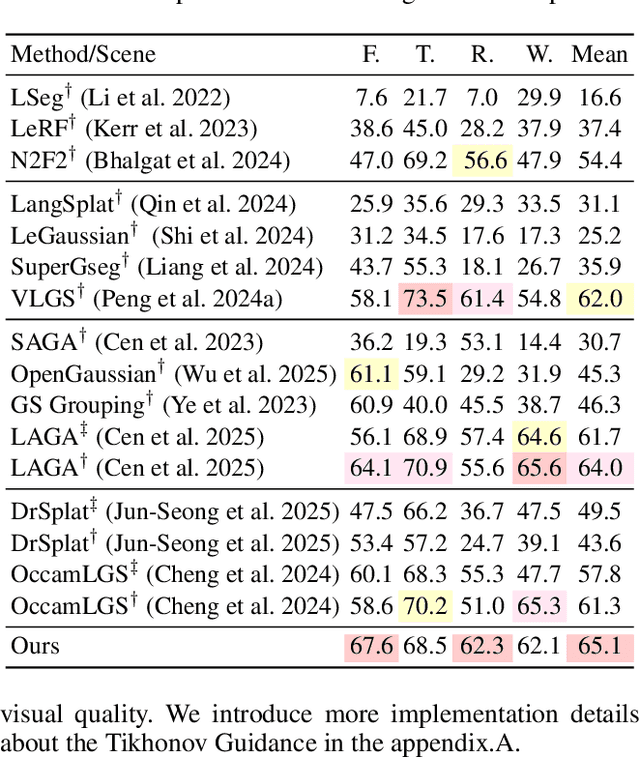
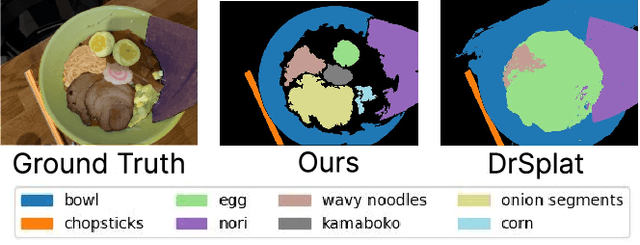
Abstract:Feature lifting has emerged as a crucial component in 3D scene understanding, enabling the attachment of rich image feature descriptors (e.g., DINO, CLIP) onto splat-based 3D representations. The core challenge lies in optimally assigning rich general attributes to 3D primitives while addressing the inconsistency issues from multi-view images. We present a unified, kernel- and feature-agnostic formulation of the feature lifting problem as a sparse linear inverse problem, which can be solved efficiently in closed form. Our approach admits a provable upper bound on the global optimal error under convex losses for delivering high quality lifted features. To address inconsistencies and noise in multi-view observations, we introduce two complementary regularization strategies to stabilize the solution and enhance semantic fidelity. Tikhonov Guidance enforces numerical stability through soft diagonal dominance, while Post-Lifting Aggregation filters noisy inputs via feature clustering. Extensive experiments demonstrate that our approach achieves state-of-the-art performance on open-vocabulary 3D segmentation benchmarks, outperforming training-based, grouping-based, and heuristic-forward baselines while producing the lifted features in minutes. Code is available at \href{https://github.com/saliteta/splat-distiller.git}{\textbf{github}}. We also have a \href{https://splat-distiller.pages.dev/}
PromptGAR: Flexible Promptive Group Activity Recognition
Mar 11, 2025



Abstract:We present PromptGAR, a novel framework that addresses the limitations of current Group Activity Recognition (GAR) approaches by leveraging multi-modal prompts to achieve both input flexibility and high recognition accuracy. The existing approaches suffer from limited real-world applicability due to their reliance on full prompt annotations, the lack of long-term actor consistency, and under-exploration of multi-group scenarios. To bridge the gap, we proposed PromptGAR, which is the first GAR model to provide input flexibility across prompts, frames, and instances without the need for retraining. Specifically, we unify bounding boxes, skeletal keypoints, and areas as point prompts and employ a recognition decoder for cross-updating class and prompt tokens. To ensure long-term consistency for extended activity durations, we also introduce a relative instance attention mechanism that directly encodes instance IDs. Finally, PromptGAR explores the use of area prompts to enable the selective recognition of the particular group activity within videos that contain multiple concurrent groups. Comprehensive evaluations demonstrate that PromptGAR achieves competitive performances both on full prompts and diverse prompt inputs, establishing its effectiveness on input flexibility and generalization ability for real-world applications.
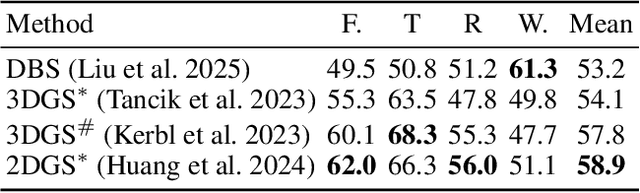
Deformable Beta Splatting
Jan 27, 2025



Abstract:3D Gaussian Splatting (3DGS) has advanced radiance field reconstruction by enabling real-time rendering. However, its reliance on Gaussian kernels for geometry and low-order Spherical Harmonics (SH) for color encoding limits its ability to capture complex geometries and diverse colors. We introduce Deformable Beta Splatting (DBS), a deformable and compact approach that enhances both geometry and color representation. DBS replaces Gaussian kernels with deformable Beta Kernels, which offer bounded support and adaptive frequency control to capture fine geometric details with higher fidelity while achieving better memory efficiency. In addition, we extended the Beta Kernel to color encoding, which facilitates improved representation of diffuse and specular components, yielding superior results compared to SH-based methods. Furthermore, Unlike prior densification techniques that depend on Gaussian properties, we mathematically prove that adjusting regularized opacity alone ensures distribution-preserved Markov chain Monte Carlo (MCMC), independent of the splatting kernel type. Experimental results demonstrate that DBS achieves state-of-the-art visual quality while utilizing only 45% of the parameters and rendering 1.5x faster than 3DGS-based methods. Notably, for the first time, splatting-based methods outperform state-of-the-art Neural Radiance Fields, highlighting the superior performance and efficiency of DBS for real-time radiance field rendering.
SplatMAP: Online Dense Monocular SLAM with 3D Gaussian Splatting
Jan 14, 2025
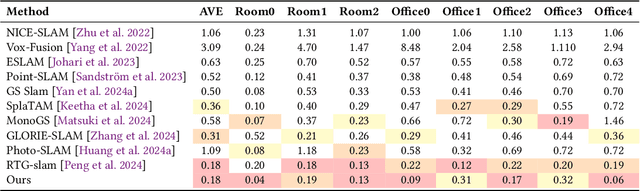

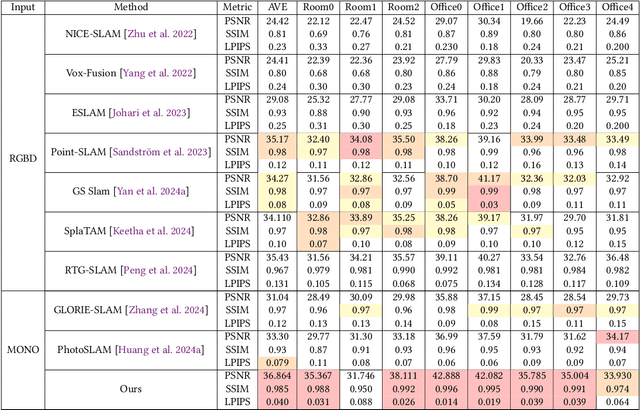
Abstract:Achieving high-fidelity 3D reconstruction from monocular video remains challenging due to the inherent limitations of traditional methods like Structure-from-Motion (SfM) and monocular SLAM in accurately capturing scene details. While differentiable rendering techniques such as Neural Radiance Fields (NeRF) address some of these challenges, their high computational costs make them unsuitable for real-time applications. Additionally, existing 3D Gaussian Splatting (3DGS) methods often focus on photometric consistency, neglecting geometric accuracy and failing to exploit SLAM's dynamic depth and pose updates for scene refinement. We propose a framework integrating dense SLAM with 3DGS for real-time, high-fidelity dense reconstruction. Our approach introduces SLAM-Informed Adaptive Densification, which dynamically updates and densifies the Gaussian model by leveraging dense point clouds from SLAM. Additionally, we incorporate Geometry-Guided Optimization, which combines edge-aware geometric constraints and photometric consistency to jointly optimize the appearance and geometry of the 3DGS scene representation, enabling detailed and accurate SLAM mapping reconstruction. Experiments on the Replica and TUM-RGBD datasets demonstrate the effectiveness of our approach, achieving state-of-the-art results among monocular systems. Specifically, our method achieves a PSNR of 36.864, SSIM of 0.985, and LPIPS of 0.040 on Replica, representing improvements of 10.7%, 6.4%, and 49.4%, respectively, over the previous SOTA. On TUM-RGBD, our method outperforms the closest baseline by 10.2%, 6.6%, and 34.7% in the same metrics. These results highlight the potential of our framework in bridging the gap between photometric and geometric dense 3D scene representations, paving the way for practical and efficient monocular dense reconstruction.
C-FedRAG: A Confidential Federated Retrieval-Augmented Generation System
Dec 17, 2024



Abstract:Organizations seeking to utilize Large Language Models (LLMs) for knowledge querying and analysis often encounter challenges in maintaining an LLM fine-tuned on targeted, up-to-date information that keeps answers relevant and grounded. Retrieval Augmented Generation (RAG) has quickly become a feasible solution for organizations looking to overcome the challenges of maintaining proprietary models and to help reduce LLM hallucinations in their query responses. However, RAG comes with its own issues regarding scaling data pipelines across tiered-access and disparate data sources. In many scenarios, it is necessary to query beyond a single data silo to provide richer and more relevant context for an LLM. Analyzing data sources within and across organizational trust boundaries is often limited by complex data-sharing policies that prohibit centralized data storage, therefore, inhibit the fast and effective setup and scaling of RAG solutions. In this paper, we introduce Confidential Computing (CC) techniques as a solution for secure Federated Retrieval Augmented Generation (FedRAG). Our proposed Confidential FedRAG system (C-FedRAG) enables secure connection and scaling of a RAG workflows across a decentralized network of data providers by ensuring context confidentiality. We also demonstrate how to implement a C-FedRAG system using the NVIDIA FLARE SDK and assess its performance using the MedRAG toolkit and MIRAGE benchmarking dataset.
Open-Vocabulary High-Resolution 3D (OVHR3D) Data Segmentation and Annotation Framework
Dec 09, 2024Abstract:In the domain of the U.S. Army modeling and simulation, the availability of high quality annotated 3D data is pivotal to creating virtual environments for training and simulations. Traditional methodologies for 3D semantic and instance segmentation, such as KpConv, RandLA, Mask3D, etc., are designed to train on extensive labeled datasets to obtain satisfactory performance in practical tasks. This requirement presents a significant challenge, given the inherent scarcity of manually annotated 3D datasets, particularly for the military use cases. Recognizing this gap, our previous research leverages the One World Terrain data repository manually annotated databases, as showcased at IITSEC 2019 and 2021, to enrich the training dataset for deep learning models. However, collecting and annotating large scale 3D data for specific tasks remains costly and inefficient. To this end, the objective of this research is to design and develop a comprehensive and efficient framework for 3D segmentation tasks to assist in 3D data annotation. This framework integrates Grounding DINO and Segment anything Model, augmented by an enhancement in 2D image rendering via 3D mesh. Furthermore, the authors have also developed a user friendly interface that facilitates the 3D annotation process, offering intuitive visualization of rendered images and the 3D point cloud.
Skyeyes: Ground Roaming using Aerial View Images
Sep 25, 2024

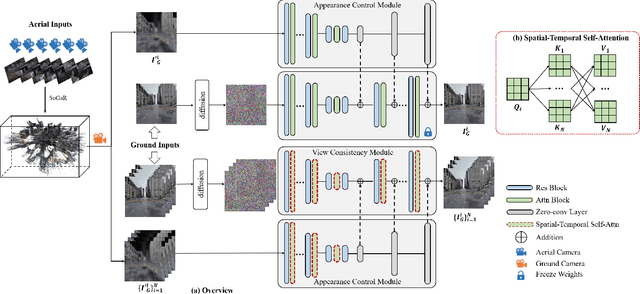

Abstract:Integrating aerial imagery-based scene generation into applications like autonomous driving and gaming enhances realism in 3D environments, but challenges remain in creating detailed content for occluded areas and ensuring real-time, consistent rendering. In this paper, we introduce Skyeyes, a novel framework that can generate photorealistic sequences of ground view images using only aerial view inputs, thereby creating a ground roaming experience. More specifically, we combine a 3D representation with a view consistent generation model, which ensures coherence between generated images. This method allows for the creation of geometrically consistent ground view images, even with large view gaps. The images maintain improved spatial-temporal coherence and realism, enhancing scene comprehension and visualization from aerial perspectives. To the best of our knowledge, there are no publicly available datasets that contain pairwise geo-aligned aerial and ground view imagery. Therefore, we build a large, synthetic, and geo-aligned dataset using Unreal Engine. Both qualitative and quantitative analyses on this synthetic dataset display superior results compared to other leading synthesis approaches. See the project page for more results: https://chaoren2357.github.io/website-skyeyes/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge