Amil Dravid
Vision Transformers Don't Need Trained Registers
Jun 09, 2025



Abstract:We investigate the mechanism underlying a previously identified phenomenon in Vision Transformers -- the emergence of high-norm tokens that lead to noisy attention maps. We observe that in multiple models (e.g., CLIP, DINOv2), a sparse set of neurons is responsible for concentrating high-norm activations on outlier tokens, leading to irregular attention patterns and degrading downstream visual processing. While the existing solution for removing these outliers involves retraining models from scratch with additional learned register tokens, we use our findings to create a training-free approach to mitigate these artifacts. By shifting the high-norm activations from our discovered register neurons into an additional untrained token, we can mimic the effect of register tokens on a model already trained without registers. We demonstrate that our method produces cleaner attention and feature maps, enhances performance over base models across multiple downstream visual tasks, and achieves results comparable to models explicitly trained with register tokens. We then extend test-time registers to off-the-shelf vision-language models to improve their interpretability. Our results suggest that test-time registers effectively take on the role of register tokens at test-time, offering a training-free solution for any pre-trained model released without them.
Interpreting the Weight Space of Customized Diffusion Models
Jun 13, 2024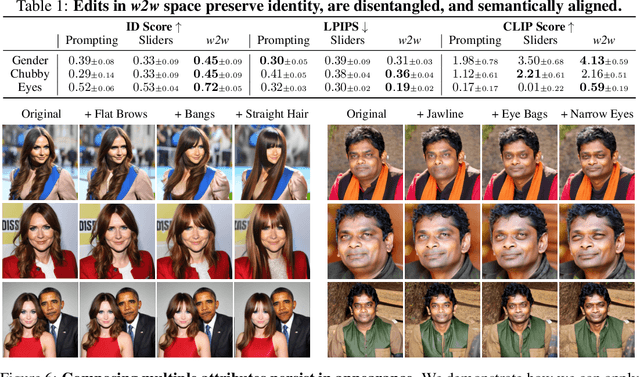


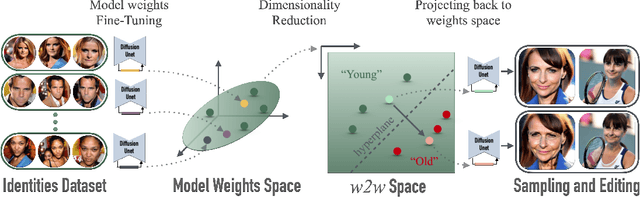
Abstract:We investigate the space of weights spanned by a large collection of customized diffusion models. We populate this space by creating a dataset of over 60,000 models, each of which is a base model fine-tuned to insert a different person's visual identity. We model the underlying manifold of these weights as a subspace, which we term weights2weights. We demonstrate three immediate applications of this space -- sampling, editing, and inversion. First, as each point in the space corresponds to an identity, sampling a set of weights from it results in a model encoding a novel identity. Next, we find linear directions in this space corresponding to semantic edits of the identity (e.g., adding a beard). These edits persist in appearance across generated samples. Finally, we show that inverting a single image into this space reconstructs a realistic identity, even if the input image is out of distribution (e.g., a painting). Our results indicate that the weight space of fine-tuned diffusion models behaves as an interpretable latent space of identities.
Idempotent Generative Network
Nov 02, 2023



Abstract:We propose a new approach for generative modeling based on training a neural network to be idempotent. An idempotent operator is one that can be applied sequentially without changing the result beyond the initial application, namely $f(f(z))=f(z)$. The proposed model $f$ is trained to map a source distribution (e.g, Gaussian noise) to a target distribution (e.g. realistic images) using the following objectives: (1) Instances from the target distribution should map to themselves, namely $f(x)=x$. We define the target manifold as the set of all instances that $f$ maps to themselves. (2) Instances that form the source distribution should map onto the defined target manifold. This is achieved by optimizing the idempotence term, $f(f(z))=f(z)$ which encourages the range of $f(z)$ to be on the target manifold. Under ideal assumptions such a process provably converges to the target distribution. This strategy results in a model capable of generating an output in one step, maintaining a consistent latent space, while also allowing sequential applications for refinement. Additionally, we find that by processing inputs from both target and source distributions, the model adeptly projects corrupted or modified data back to the target manifold. This work is a first step towards a ``global projector'' that enables projecting any input into a target data distribution.
Rosetta Neurons: Mining the Common Units in a Model Zoo
Jun 16, 2023Abstract:Do different neural networks, trained for various vision tasks, share some common representations? In this paper, we demonstrate the existence of common features we call "Rosetta Neurons" across a range of models with different architectures, different tasks (generative and discriminative), and different types of supervision (class-supervised, text-supervised, self-supervised). We present an algorithm for mining a dictionary of Rosetta Neurons across several popular vision models: Class Supervised-ResNet50, DINO-ResNet50, DINO-ViT, MAE, CLIP-ResNet50, BigGAN, StyleGAN-2, StyleGAN-XL. Our findings suggest that certain visual concepts and structures are inherently embedded in the natural world and can be learned by different models regardless of the specific task or architecture, and without the use of semantic labels. We can visualize shared concepts directly due to generative models included in our analysis. The Rosetta Neurons facilitate model-to-model translation enabling various inversion-based manipulations, including cross-class alignments, shifting, zooming, and more, without the need for specialized training.
DeepCOVID-Fuse: A Multi-modality Deep Learning Model Fusing Chest X-Radiographs and Clinical Variables to Predict COVID-19 Risk Levels
Jan 20, 2023Abstract:Propose: To present DeepCOVID-Fuse, a deep learning fusion model to predict risk levels in patients with confirmed coronavirus disease 2019 (COVID-19) and to evaluate the performance of pre-trained fusion models on full or partial combination of chest x-ray (CXRs) or chest radiograph and clinical variables. Materials and Methods: The initial CXRs, clinical variables and outcomes (i.e., mortality, intubation, hospital length of stay, ICU admission) were collected from February 2020 to April 2020 with reverse-transcription polymerase chain reaction (RT-PCR) test results as the reference standard. The risk level was determined by the outcome. The fusion model was trained on 1657 patients (Age: 58.30 +/- 17.74; Female: 807) and validated on 428 patients (56.41 +/- 17.03; 190) from Northwestern Memorial HealthCare system and was tested on 439 patients (56.51 +/- 17.78; 205) from a single holdout hospital. Performance of pre-trained fusion models on full or partial modalities were compared on the test set using the DeLong test for the area under the receiver operating characteristic curve (AUC) and the McNemar test for accuracy, precision, recall and F1. Results: The accuracy of DeepCOVID-Fuse trained on CXRs and clinical variables is 0.658, with an AUC of 0.842, which significantly outperformed (p < 0.05) models trained only on CXRs with an accuracy of 0.621 and AUC of 0.807 and only on clinical variables with an accuracy of 0.440 and AUC of 0.502. The pre-trained fusion model with only CXRs as input increases accuracy to 0.632 and AUC to 0.813 and with only clinical variables as input increases accuracy to 0.539 and AUC to 0.733. Conclusion: The fusion model learns better feature representations across different modalities during training and achieves good outcome predictions even when only some of the modalities are used in testing.
BKinD-3D: Self-Supervised 3D Keypoint Discovery from Multi-View Videos
Dec 14, 2022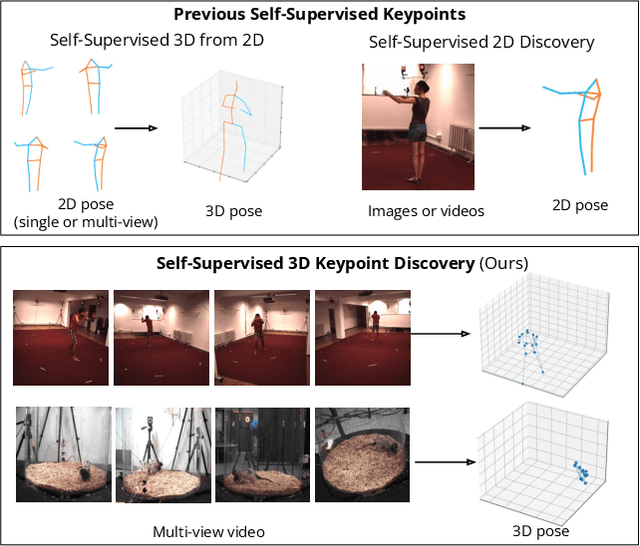
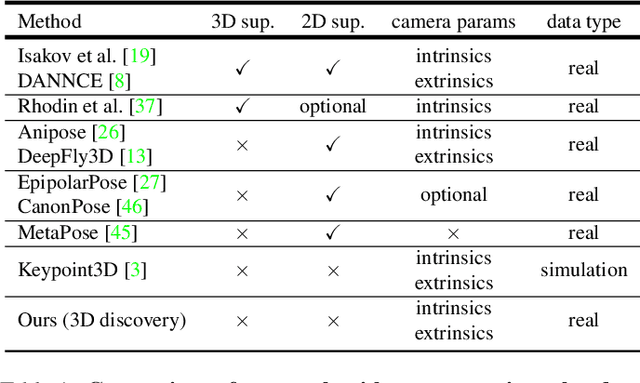
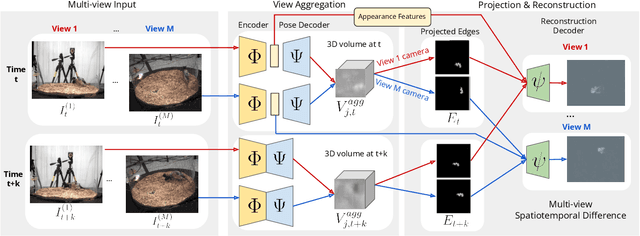
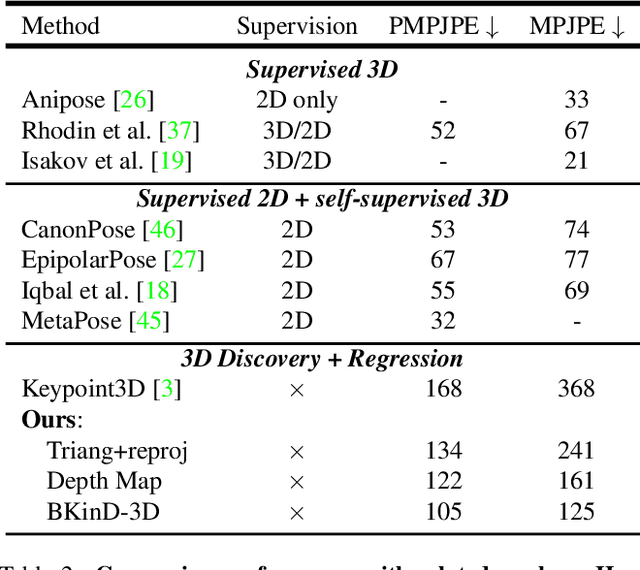
Abstract:Quantifying motion in 3D is important for studying the behavior of humans and other animals, but manual pose annotations are expensive and time-consuming to obtain. Self-supervised keypoint discovery is a promising strategy for estimating 3D poses without annotations. However, current keypoint discovery approaches commonly process single 2D views and do not operate in the 3D space. We propose a new method to perform self-supervised keypoint discovery in 3D from multi-view videos of behaving agents, without any keypoint or bounding box supervision in 2D or 3D. Our method uses an encoder-decoder architecture with a 3D volumetric heatmap, trained to reconstruct spatiotemporal differences across multiple views, in addition to joint length constraints on a learned 3D skeleton of the subject. In this way, we discover keypoints without requiring manual supervision in videos of humans and rats, demonstrating the potential of 3D keypoint discovery for studying behavior.
medXGAN: Visual Explanations for Medical Classifiers through a Generative Latent Space
Apr 17, 2022



Abstract:Despite the surge of deep learning in the past decade, some users are skeptical to deploy these models in practice due to their black-box nature. Specifically, in the medical space where there are severe potential repercussions, we need to develop methods to gain confidence in the models' decisions. To this end, we propose a novel medical imaging generative adversarial framework, medXGAN (medical eXplanation GAN), to visually explain what a medical classifier focuses on in its binary predictions. By encoding domain knowledge of medical images, we are able to disentangle anatomical structure and pathology, leading to fine-grained visualization through latent interpolation. Furthermore, we optimize the latent space such that interpolation explains how the features contribute to the classifier's output. Our method outperforms baselines such as Gradient-Weighted Class Activation Mapping (Grad-CAM) and Integrated Gradients in localization and explanatory ability. Additionally, a combination of the medXGAN with Integrated Gradients can yield explanations more robust to noise. The code is available at: https://avdravid.github.io/medXGAN_page/.
Investigating the Potential of Auxiliary-Classifier GANs for Image Classification in Low Data Regimes
Jan 22, 2022



Abstract:Generative Adversarial Networks (GANs) have shown promise in augmenting datasets and boosting convolutional neural networks' (CNN) performance on image classification tasks. But they introduce more hyperparameters to tune as well as the need for additional time and computational power to train supplementary to the CNN. In this work, we examine the potential for Auxiliary-Classifier GANs (AC-GANs) as a 'one-stop-shop' architecture for image classification, particularly in low data regimes. Additionally, we explore modifications to the typical AC-GAN framework, changing the generator's latent space sampling scheme and employing a Wasserstein loss with gradient penalty to stabilize the simultaneous training of image synthesis and classification. Through experiments on images of varying resolutions and complexity, we demonstrate that AC-GANs show promise in image classification, achieving competitive performance with standard CNNs. These methods can be employed as an 'all-in-one' framework with particular utility in the absence of large amounts of training data.
Visual Explanations for Convolutional Neural Networks via Latent Traversal of Generative Adversarial Networks
Nov 02, 2021

Abstract:Lack of explainability in artificial intelligence, specifically deep neural networks, remains a bottleneck for implementing models in practice. Popular techniques such as Gradient-weighted Class Activation Mapping (Grad-CAM) provide a coarse map of salient features in an image, which rarely tells the whole story of what a convolutional neural network (CNN) learned. Using COVID-19 chest X-rays, we present a method for interpreting what a CNN has learned by utilizing Generative Adversarial Networks (GANs). Our GAN framework disentangles lung structure from COVID-19 features. Using this GAN, we can visualize the transition of a pair of COVID negative lungs in a chest radiograph to a COVID positive pair by interpolating in the latent space of the GAN, which provides fine-grained visualization of how the CNN responds to varying features within the lungs.
Interpretation of Brain Morphology in Association to Alzheimer's Disease Dementia Classification Using Graph Convolutional Networks on Triangulated Meshes
Aug 20, 2020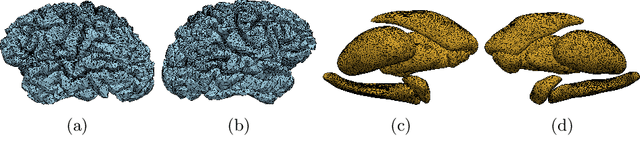
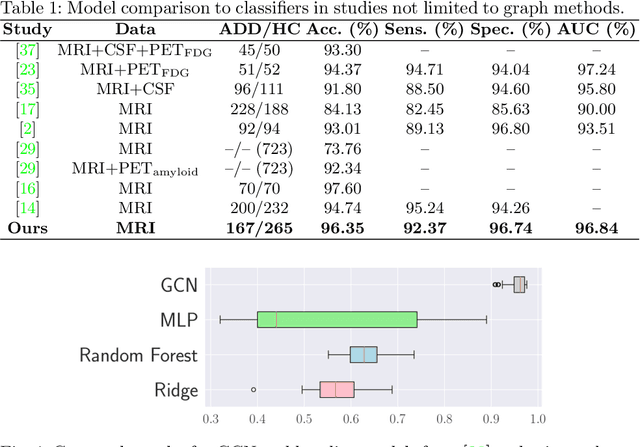
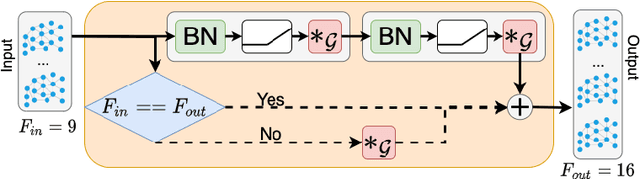
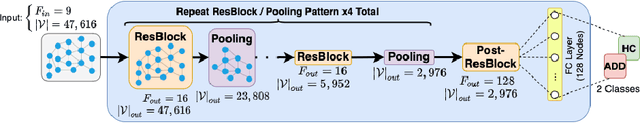
Abstract:We propose a mesh-based technique to aid in the classification of Alzheimer's disease dementia (ADD) using mesh representations of the cortex and subcortical structures. Deep learning methods for classification tasks that utilize structural neuroimaging often require extensive learning parameters to optimize. Frequently, these approaches for automated medical diagnosis also lack visual interpretability for areas in the brain involved in making a diagnosis. This work: (a) analyzes brain shape using surface information of the cortex and subcortical structures, (b) proposes a residual learning framework for state-of-the-art graph convolutional networks which offer a significant reduction in learnable parameters, and (c) offers visual interpretability of the network via class-specific gradient information that localizes important regions of interest in our inputs. With our proposed method leveraging the use of cortical and subcortical surface information, we outperform other machine learning methods with a 96.35% testing accuracy for the ADD vs. healthy control problem. We confirm the validity of our model by observing its performance in a 25-trial Monte Carlo cross-validation. The generated visualization maps in our study show correspondences with current knowledge regarding the structural localization of pathological changes in the brain associated to dementia of the Alzheimer's type.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge