Adrian Florea
XCOMPS: A Multilingual Benchmark of Conceptual Minimal Pairs
Feb 27, 2025



Abstract:We introduce XCOMPS in this work, a multilingual conceptual minimal pair dataset covering 17 languages. Using this dataset, we evaluate LLMs' multilingual conceptual understanding through metalinguistic prompting, direct probability measurement, and neurolinguistic probing. By comparing base, instruction-tuned, and knowledge-distilled models, we find that: 1) LLMs exhibit weaker conceptual understanding for low-resource languages, and accuracy varies across languages despite being tested on the same concept sets. 2) LLMs excel at distinguishing concept-property pairs that are visibly different but exhibit a marked performance drop when negative pairs share subtle semantic similarities. 3) Instruction tuning improves performance in concept understanding but does not enhance internal competence; knowledge distillation can enhance internal competence in conceptual understanding for low-resource languages with limited gains in explicit task performance. 4) More morphologically complex languages yield lower concept understanding scores and require deeper layers for conceptual reasoning.
Exploring Finetuned Audio-LLM on Heart Murmur Features
Jan 23, 2025



Abstract:Large language models (LLMs) for audio have excelled in recognizing and analyzing human speech, music, and environmental sounds. However, their potential for understanding other types of sounds, particularly biomedical sounds, remains largely underexplored despite significant scientific interest. In this study, we focus on diagnosing cardiovascular diseases using phonocardiograms, i.e., heart sounds. Most existing deep neural network (DNN) paradigms are restricted to heart murmur classification (healthy vs unhealthy) and do not predict other acoustic features of the murmur such as timing, grading, harshness, pitch, and quality, which are important in helping physicians diagnose the underlying heart conditions. We propose to finetune an audio LLM, Qwen2-Audio, on the PhysioNet CirCor DigiScope phonocardiogram (PCG) dataset and evaluate its performance in classifying 11 expert-labeled murmur features. Additionally, we aim to achieve more noise-robust and generalizable system by exploring a preprocessing segmentation algorithm using an audio representation model, SSAMBA. Our results indicate that the LLM-based model outperforms state-of-the-art methods in 8 of the 11 features and performs comparably in the remaining 3. Moreover, the LLM successfully classifies long-tail murmur features with limited training data, a task that all previous methods have failed to classify. These findings underscore the potential of audio LLMs as assistants to human cardiologists in enhancing heart disease diagnosis.
Estimating Rural Path Loss with ITU-R P.1812-7 : Impact of Geospatial Inputs
Jan 20, 2025Abstract:Accurate radio wave propagation modeling is essential for effective spectrum management by regulators and network deployment by operators. This paper investigates the ITU-R P.1812-7 (P.1812) propagation model's reliance on geospatial inputs, particularly clutter information, to improve path loss estimation, with an emphasis on rural geographic regions. The research evaluates the impact of geospatial elevation and land cover datasets, including Global Forest Canopy Height (GFCH), European Space Agency WorldCover, and Natural Resources Canada LandCover, on P.1812 propagation model prediction accuracy. Results highlight the trade-offs between dataset resolution, geospatial data availability, and representative clutter height assignments. Simulations reveal that high-resolution data do not always yield better results and that global datasets such as the GFCH provide a robust alternative when high-resolution data are unavailable or out-of-date. This study provides a set of guidelines for geospatial dataset integration to enhance P.1812's rural path loss predictions.
COVID-Net USPro: An Open-Source Explainable Few-Shot Deep Prototypical Network to Monitor and Detect COVID-19 Infection from Point-of-Care Ultrasound Images
Jan 04, 2023Abstract:As the Coronavirus Disease 2019 (COVID-19) continues to impact many aspects of life and the global healthcare systems, the adoption of rapid and effective screening methods to prevent further spread of the virus and lessen the burden on healthcare providers is a necessity. As a cheap and widely accessible medical image modality, point-of-care ultrasound (POCUS) imaging allows radiologists to identify symptoms and assess severity through visual inspection of the chest ultrasound images. Combined with the recent advancements in computer science, applications of deep learning techniques in medical image analysis have shown promising results, demonstrating that artificial intelligence-based solutions can accelerate the diagnosis of COVID-19 and lower the burden on healthcare professionals. However, the lack of a huge amount of well-annotated data poses a challenge in building effective deep neural networks in the case of novel diseases and pandemics. Motivated by this, we present COVID-Net USPro, an explainable few-shot deep prototypical network, that monitors and detects COVID-19 positive cases with high precision and recall from minimal ultrasound images. COVID-Net USPro achieves 99.65% overall accuracy, 99.7% recall and 99.67% precision for COVID-19 positive cases when trained with only 5 shots. The analytic pipeline and results were verified by our contributing clinician with extensive experience in POCUS interpretation, ensuring that the network makes decisions based on actual patterns.
COVID-Net US-X: Enhanced Deep Neural Network for Detection of COVID-19 Patient Cases from Convex Ultrasound Imaging Through Extended Linear-Convex Ultrasound Augmentation Learning
Apr 29, 2022



Abstract:As the global population continues to face significant negative impact by the on-going COVID-19 pandemic, there has been an increasing usage of point-of-care ultrasound (POCUS) imaging as a low-cost and effective imaging modality of choice in the COVID-19 clinical workflow. A major barrier with widespread adoption of POCUS in the COVID-19 clinical workflow is the scarcity of expert clinicians that can interpret POCUS examinations, leading to considerable interest in deep learning-driven clinical decision support systems to tackle this challenge. A major challenge to building deep neural networks for COVID-19 screening using POCUS is the heterogeneity in the types of probes used to capture ultrasound images (e.g., convex vs. linear probes), which can lead to very different visual appearances. In this study, we explore the impact of leveraging extended linear-convex ultrasound augmentation learning on producing enhanced deep neural networks for COVID-19 assessment, where we conduct data augmentation on convex probe data alongside linear probe data that have been transformed to better resemble convex probe data. Experimental results using an efficient deep columnar anti-aliased convolutional neural network designed via a machined-driven design exploration strategy (which we name COVID-Net US-X) show that the proposed extended linear-convex ultrasound augmentation learning significantly increases performance, with a gain of 5.1% in test accuracy and 13.6% in AUC.
COVID-Net Biochem: An Explainability-driven Framework to Building Machine Learning Models for Predicting Survival and Kidney Injury of COVID-19 Patients from Clinical and Biochemistry Data
Apr 24, 2022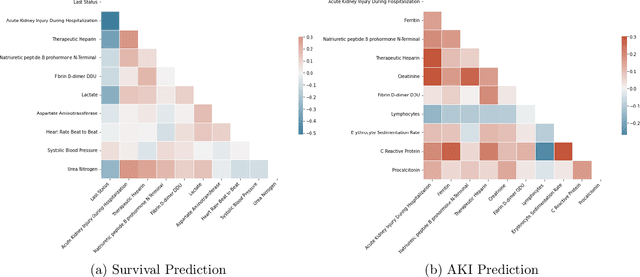
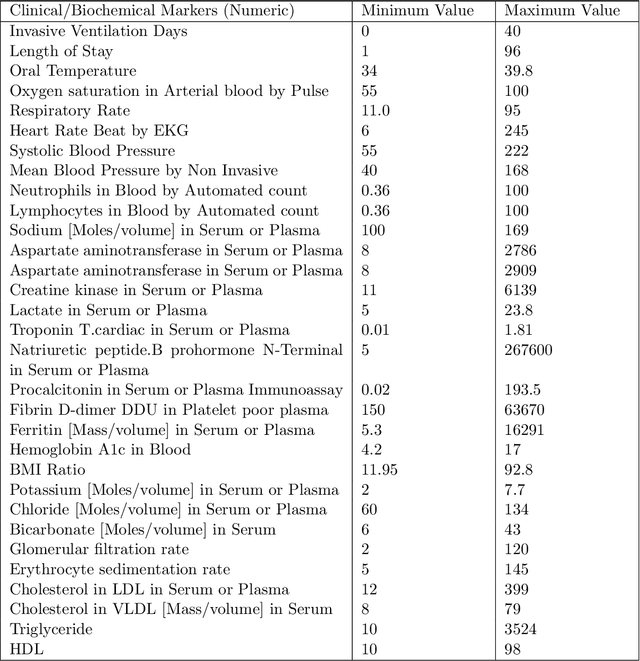
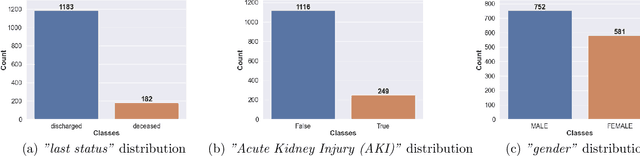
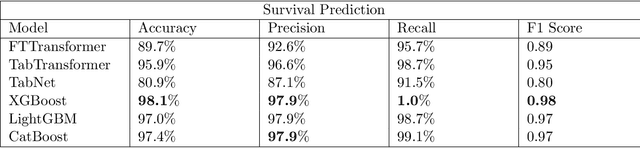
Abstract:Ever since the declaration of COVID-19 as a pandemic by the World Health Organization in 2020, the world has continued to struggle in controlling and containing the spread of the COVID-19 pandemic caused by the SARS-CoV-2 virus. This has been especially challenging with the rise of the Omicron variant and its subvariants and recombinants, which has led to a significant increase in patients seeking treatment and has put a tremendous burden on hospitals and healthcare systems. A major challenge faced during the pandemic has been the prediction of survival and the risk for additional injuries in individual patients, which requires significant clinical expertise and additional resources to avoid further complications. In this study we propose COVID-Net Biochem, an explainability-driven framework for building machine learning models to predict patient survival and the chance of developing kidney injury during hospitalization from clinical and biochemistry data in a transparent and systematic manner. In the first "clinician-guided initial design" phase, we prepared a benchmark dataset of carefully selected clinical and biochemistry data based on clinician assessment, which were curated from a patient cohort of 1366 patients at Stony Brook University. A collection of different machine learning models with a diversity of gradient based boosting tree architectures and deep transformer architectures was designed and trained specifically for survival and kidney injury prediction based on the carefully selected clinical and biochemical markers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge