Abhyuday Jagannatha
University of Massachusetts Amherst
Object-Aware Cropping for Self-Supervised Learning
Dec 01, 2021



Abstract:A core component of the recent success of self-supervised learning is cropping data augmentation, which selects sub-regions of an image to be used as positive views in the self-supervised loss. The underlying assumption is that randomly cropped and resized regions of a given image share information about the objects of interest, which the learned representation will capture. This assumption is mostly satisfied in datasets such as ImageNet where there is a large, centered object, which is highly likely to be present in random crops of the full image. However, in other datasets such as OpenImages or COCO, which are more representative of real world uncurated data, there are typically multiple small objects in an image. In this work, we show that self-supervised learning based on the usual random cropping performs poorly on such datasets. We propose replacing one or both of the random crops with crops obtained from an object proposal algorithm. This encourages the model to learn both object and scene level semantic representations. Using this approach, which we call object-aware cropping, results in significant improvements over scene cropping on classification and object detection benchmarks. For example, on OpenImages, our approach achieves an improvement of 8.8% mAP over random scene-level cropping using MoCo-v2 based pre-training. We also show significant improvements on COCO and PASCAL-VOC object detection and segmentation tasks over the state-of-the-art self-supervised learning approaches. Our approach is efficient, simple and general, and can be used in most existing contrastive and non-contrastive self-supervised learning frameworks.
Membership Inference Attack Susceptibility of Clinical Language Models
Apr 16, 2021



Abstract:Deep Neural Network (DNN) models have been shown to have high empirical privacy leakages. Clinical language models (CLMs) trained on clinical data have been used to improve performance in biomedical natural language processing tasks. In this work, we investigate the risks of training-data leakage through white-box or black-box access to CLMs. We design and employ membership inference attacks to estimate the empirical privacy leaks for model architectures like BERT and GPT2. We show that membership inference attacks on CLMs lead to non-trivial privacy leakages of up to 7%. Our results show that smaller models have lower empirical privacy leakages than larger ones, and masked LMs have lower leakages than auto-regressive LMs. We further show that differentially private CLMs can have improved model utility on clinical domain while ensuring low empirical privacy leakage. Lastly, we also study the effects of group-level membership inference and disease rarity on CLM privacy leakages.
Federated Intrusion Detection for IoT with Heterogeneous Cohort Privacy
Jan 25, 2021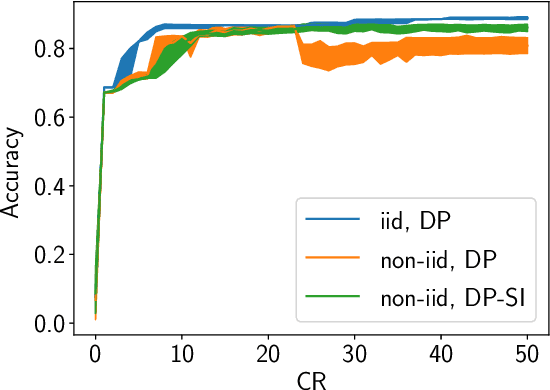

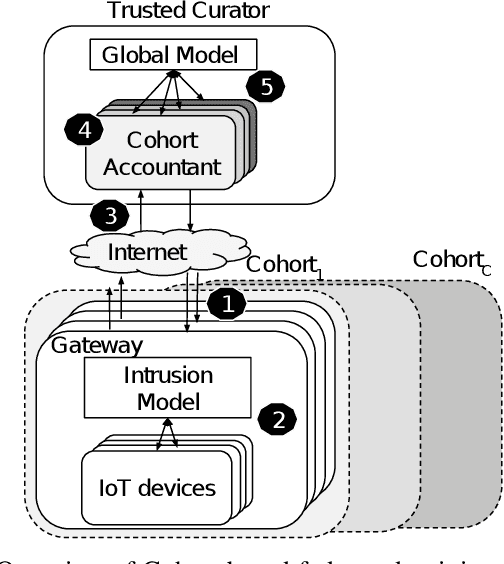
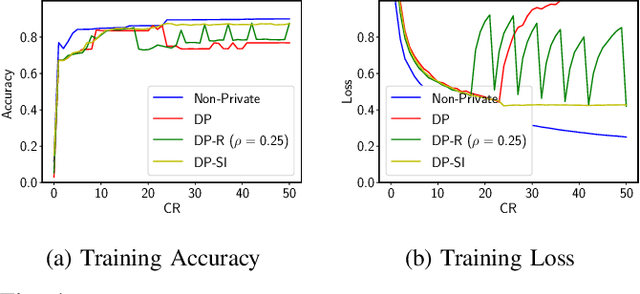
Abstract:Internet of Things (IoT) devices are becoming increasingly popular and are influencing many application domains such as healthcare and transportation. These devices are used for real-world applications such as sensor monitoring, real-time control. In this work, we look at differentially private (DP) neural network (NN) based network intrusion detection systems (NIDS) to detect intrusion attacks on networks of such IoT devices. Existing NN training solutions in this domain either ignore privacy considerations or assume that the privacy requirements are homogeneous across all users. We show that the performance of existing differentially private stochastic methods degrade for clients with non-identical data distributions when clients' privacy requirements are heterogeneous. We define a cohort-based $(\epsilon,\delta)$-DP framework that models the more practical setting of IoT device cohorts with non-identical clients and heterogeneous privacy requirements. We propose two novel continual-learning based DP training methods that are designed to improve model performance in the aforementioned setting. To the best of our knowledge, ours is the first system that employs a continual learning-based approach to handle heterogeneity in client privacy requirements. We evaluate our approach on real datasets and show that our techniques outperform the baselines. We also show that our methods are robust to hyperparameter changes. Lastly, we show that one of our proposed methods can easily adapt to post-hoc relaxations of client privacy requirements.
Calibrating Structured Output Predictors for Natural Language Processing
May 05, 2020



Abstract:We address the problem of calibrating prediction confidence for output entities of interest in natural language processing (NLP) applications. It is important that NLP applications such as named entity recognition and question answering produce calibrated confidence scores for their predictions, especially if the system is to be deployed in a safety-critical domain such as healthcare. However, the output space of such structured prediction models is often too large to adapt binary or multi-class calibration methods directly. In this study, we propose a general calibration scheme for output entities of interest in neural-network based structured prediction models. Our proposed method can be used with any binary class calibration scheme and a neural network model. Additionally, we show that our calibration method can also be used as an uncertainty-aware, entity-specific decoding step to improve the performance of the underlying model at no additional training cost or data requirements. We show that our method outperforms current calibration techniques for named-entity-recognition, part-of-speech and question answering. We also improve our model's performance from our decoding step across several tasks and benchmark datasets. Our method improves the calibration and model performance on out-of-domain test scenarios as well.
Towards Automatic Generation of Questions from Long Answers
Apr 15, 2020



Abstract:Automatic question generation (AQG) has broad applicability in domains such as tutoring systems, conversational agents, healthcare literacy, and information retrieval. Existing efforts at AQG have been limited to short answer lengths of up to two or three sentences. However, several real-world applications require question generation from answers that span several sentences. Therefore, we propose a novel evaluation benchmark to assess the performance of existing AQG systems for long-text answers. We leverage the large-scale open-source Google Natural Questions dataset to create the aforementioned long-answer AQG benchmark. We empirically demonstrate that the performance of existing AQG methods significantly degrades as the length of the answer increases. Transformer-based methods outperform other existing AQG methods on long answers in terms of automatic as well as human evaluation. However, we still observe degradation in the performance of our best performing models with increasing sentence length, suggesting that long answer QA is a challenging benchmark task for future research.
Improved Pretraining for Domain-specific Contextual Embedding Models
Apr 05, 2020

Abstract:We investigate methods to mitigate catastrophic forgetting during domain-specific pretraining of contextual embedding models such as BERT, DistilBERT, and RoBERTa. Recently proposed domain-specific models such as BioBERT, SciBERT and ClinicalBERT are constructed by continuing the pretraining phase on a domain-specific text corpus. Such pretraining is susceptible to catastrophic forgetting, where the model forgets some of the information learned in the general domain. We propose the use of two continual learning techniques (rehearsal and elastic weight consolidation) to improve domain-specific training. Our results show that models trained by our proposed approaches can better maintain their performance on the general domain tasks, and at the same time, outperform domain-specific baseline models on downstream domain tasks.
Active Learning for New Domains in Natural Language Understanding
Oct 03, 2018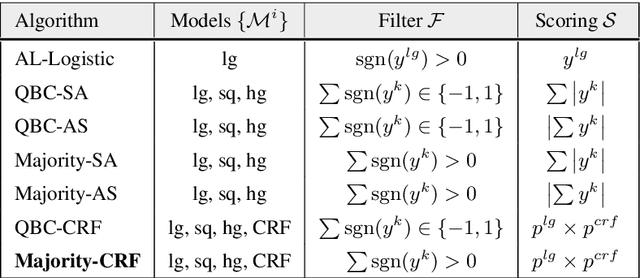
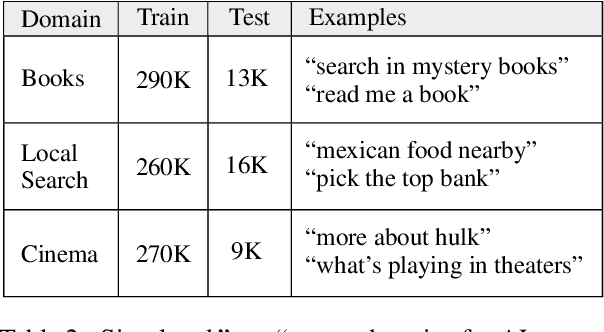
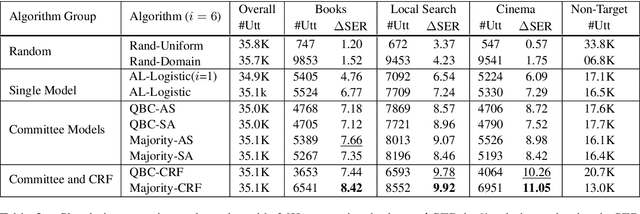
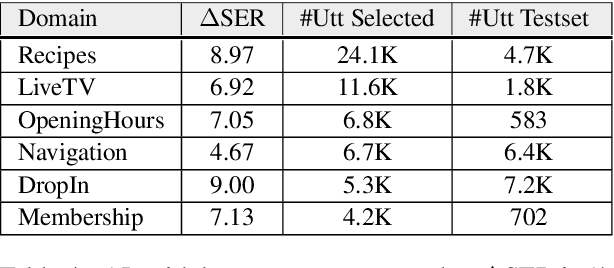
Abstract:We explore active learning (AL) utterance selection for improving the accuracy of new underrepresented domains in a natural language understanding (NLU) system. Moreover, we propose an AL algorithm called Majority-CRF that uses an ensemble of classification and sequence labeling models to guide utterance selection for annotation. Experiments with three domains show that Majority-CRF achieves 6.6%-9% relative error rate reduction compared to random sampling with the same annotation budget, and statistically significant improvements compared to other AL approaches. Additionally, case studies with human-in-the-loop AL on six new domains show 4.6%-9% improvement on an existing NLU system.
Structured prediction models for RNN based sequence labeling in clinical text
Aug 01, 2016


Abstract:Sequence labeling is a widely used method for named entity recognition and information extraction from unstructured natural language data. In clinical domain one major application of sequence labeling involves extraction of medical entities such as medication, indication, and side-effects from Electronic Health Record narratives. Sequence labeling in this domain, presents its own set of challenges and objectives. In this work we experimented with various CRF based structured learning models with Recurrent Neural Networks. We extend the previously studied LSTM-CRF models with explicit modeling of pairwise potentials. We also propose an approximate version of skip-chain CRF inference with RNN potentials. We use these methodologies for structured prediction in order to improve the exact phrase detection of various medical entities.
Bidirectional Recurrent Neural Networks for Medical Event Detection in Electronic Health Records
Jul 12, 2016



Abstract:Sequence labeling for extraction of medical events and their attributes from unstructured text in Electronic Health Record (EHR) notes is a key step towards semantic understanding of EHRs. It has important applications in health informatics including pharmacovigilance and drug surveillance. The state of the art supervised machine learning models in this domain are based on Conditional Random Fields (CRFs) with features calculated from fixed context windows. In this application, we explored various recurrent neural network frameworks and show that they significantly outperformed the CRF models.
Learning for Biomedical Information Extraction: Methodological Review of Recent Advances
Jun 26, 2016
Abstract:Biomedical information extraction (BioIE) is important to many applications, including clinical decision support, integrative biology, and pharmacovigilance, and therefore it has been an active research. Unlike existing reviews covering a holistic view on BioIE, this review focuses on mainly recent advances in learning based approaches, by systematically summarizing them into different aspects of methodological development. In addition, we dive into open information extraction and deep learning, two emerging and influential techniques and envision next generation of BioIE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge