Ptb Dataset
Papers and Code
Metric Hub: A metric library and practical selection workflow for use-case-driven data quality assessment in medical AI
Jan 30, 2026Machine learning (ML) in medicine has transitioned from research to concrete applications aimed at supporting several medical purposes like therapy selection, monitoring and treatment. Acceptance and effective adoption by clinicians and patients, as well as regulatory approval, require evidence of trustworthiness. A major factor for the development of trustworthy AI is the quantification of data quality for AI model training and testing. We have recently proposed the METRIC-framework for systematically evaluating the suitability (fit-for-purpose) of data for medical ML for a given task. Here, we operationalize this theoretical framework by introducing a collection of data quality metrics - the metric library - for practically measuring data quality dimensions. For each metric, we provide a metric card with the most important information, including definition, applicability, examples, pitfalls and recommendations, to support the understanding and implementation of these metrics. Furthermore, we discuss strategies and provide decision trees for choosing an appropriate set of data quality metrics from the metric library given specific use cases. We demonstrate the impact of our approach exemplarily on the PTB-XL ECG-dataset. This is a first step to enable fit-for-purpose evaluation of training and test data in practice as the base for establishing trustworthy AI in medicine.
How Much Temporal Modeling is Enough? A Systematic Study of Hybrid CNN-RNN Architectures for Multi-Label ECG Classification
Jan 25, 2026Accurate multi-label classification of electrocardiogram (ECG) signals remains challenging due to the coexistence of multiple cardiac conditions, pronounced class imbalance, and long-range temporal dependencies in multi-lead recordings. Although recent studies increasingly rely on deep and stacked recurrent architectures, the necessity and clinical justification of such architectural complexity have not been rigorously examined. In this work, we perform a systematic comparative evaluation of convolutional neural networks (CNNs) combined with multiple recurrent configurations, including LSTM, GRU, Bidirectional LSTM (BiLSTM), and their stacked variants, for multi-label ECG classification on the PTB-XL dataset comprising 23 diagnostic categories. The CNN component serves as a morphology-driven baseline, while recurrent layers are progressively integrated to assess their contribution to temporal modeling and generalization performance. Experimental results indicate that a CNN integrated with a single BiLSTM layer achieves the most favorable trade-off between predictive performance and model complexity. This configuration attains superior Hamming loss (0.0338), macro-AUPRC (0.4715), micro-F1 score (0.6979), and subset accuracy (0.5723) compared with deeper recurrent combinations. Although stacked recurrent models occasionally improve recall for specific rare classes, our results provide empirical evidence that increasing recurrent depth yields diminishing returns and may degrade generalization due to reduced precision and overfitting. These findings suggest that architectural alignment with the intrinsic temporal structure of ECG signals, rather than increased recurrent depth, is a key determinant of robust performance and clinically relevant deployment.
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
ECG-RAMBA: Zero-Shot ECG Generalization by Morphology-Rhythm Disentanglement and Long-Range Modeling
Dec 29, 2025Deep learning has achieved strong performance for electrocardiogram (ECG) classification within individual datasets, yet dependable generalization across heterogeneous acquisition settings remains a major obstacle to clinical deployment and longitudinal monitoring. A key limitation of many model architectures is the implicit entanglement of morphological waveform patterns and rhythm dynamics, which can promote shortcut learning and amplify sensitivity to distribution shifts. We propose ECG-RAMBA, a framework that separates morphology and rhythm and then re-integrates them through context-aware fusion. ECG-RAMBA combines: (i) deterministic morphological features extracted by MiniRocket, (ii) global rhythm descriptors computed from heart-rate variability (HRV), and (iii) long-range contextual modeling via a bi-directional Mamba backbone. To improve sensitivity to transient abnormalities under windowed inference, we introduce a numerically stable Power Mean pooling operator ($Q=3$) that emphasizes high-evidence segments while avoiding the brittleness of max pooling and the dilution of averaging. We evaluate under a protocol-faithful setting with subject-level cross-validation, a fixed decision threshold, and no test-time adaptation. On the Chapman--Shaoxing dataset, ECG-RAMBA achieves a macro ROC-AUC $\approx 0.85$. In zero-shot transfer, it attains PR-AUC $=0.708$ for atrial fibrillation detection on the external CPSC-2021 dataset, substantially outperforming a comparable raw-signal Mamba baseline, and shows consistent cross-dataset performance on PTB-XL. Ablation studies indicate that deterministic morphology provides a strong foundation, while explicit rhythm modeling and long-range context are critical drivers of cross-domain robustness.
Investigating ECG Diagnosis with Ambiguous Labels using Partial Label Learning
Dec 11, 2025Label ambiguity is an inherent problem in real-world electrocardiogram (ECG) diagnosis, arising from overlapping conditions and diagnostic disagreement. However, current ECG models are trained under the assumption of clean and non-ambiguous annotations, which limits both the development and the meaningful evaluation of models under real-world conditions. Although Partial Label Learning (PLL) frameworks are designed to learn from ambiguous labels, their effectiveness in medical time-series domains, ECG in particular, remains largely unexplored. In this work, we present the first systematic study of PLL methods for ECG diagnosis. We adapt nine PLL algorithms to multi-label ECG diagnosis and evaluate them using a diverse set of clinically motivated ambiguity generation strategies, capturing both unstructured (e.g., random) and structured ambiguities (e.g., cardiologist-derived similarities, treatment relationships, and diagnostic taxonomies). Our experiments on the PTB-XL and Chapman datasets demonstrate that PLL methods vary substantially in their robustness to different types and degrees of ambiguity. Through extensive analysis, we identify key limitations of current PLL approaches in clinical settings and outline future directions for developing robust and clinically aligned ambiguity-aware learning frameworks for ECG diagnosis.
Lightweight Multi-task CNN for ECG Diagnosis with GRU-Diffusion
Nov 18, 2025With the increasing demand for real-time Electrocardiogram (ECG) classification on edge devices, existing models face challenges of high computational cost and limited accuracy on imbalanced datasets.This paper presents Multi-task DFNet, a lightweight multi-task framework for ECG classification across the MIT-BIH Arrhythmia Database and the PTB Diagnostic ECG Database, enabling efficient task collaboration by dynamically sharing knowledge across tasks, such as arrhythmia detection, myocardial infarction (MI) classification, and other cardiovascular abnormalities. The proposed method integrates GRU-augmented Diffusion, where the GRU is embedded within the diffusion model to capture temporal dependencies better and generate high-quality synthetic signals for imbalanced classes. The experimental results show that Multi-task DFNet achieves 99.72% and 99.89% accuracy on the MIT-BIH dataset and PTB dataset, respectively, with significantly fewer parameters compared to traditional models, making it suitable for deployment on wearable ECG monitors. This work offers a compact and efficient solution for multi-task ECG diagnosis, providing a promising potential for edge healthcare applications on resource-constrained devices.
CODE-II: A large-scale dataset for artificial intelligence in ECG analysis
Nov 19, 2025Data-driven methods for electrocardiogram (ECG) interpretation are rapidly progressing. Large datasets have enabled advances in artificial intelligence (AI) based ECG analysis, yet limitations in annotation quality, size, and scope remain major challenges. Here we present CODE-II, a large-scale real-world dataset of 2,735,269 12-lead ECGs from 2,093,807 adult patients collected by the Telehealth Network of Minas Gerais (TNMG), Brazil. Each exam was annotated using standardized diagnostic criteria and reviewed by cardiologists. A defining feature of CODE-II is a set of 66 clinically meaningful diagnostic classes, developed with cardiologist input and routinely used in telehealth practice. We additionally provide an open available subset: CODE-II-open, a public subset of 15,000 patients, and the CODE-II-test, a non-overlapping set of 8,475 exams reviewed by multiple cardiologists for blinded evaluation. A neural network pre-trained on CODE-II achieved superior transfer performance on external benchmarks (PTB-XL and CPSC 2018) and outperformed alternatives trained on larger datasets.
Enhancing ECG Classification Robustness with Lightweight Unsupervised Anomaly Detection Filters
Oct 30, 2025Continuous electrocardiogram (ECG) monitoring via wearables offers significant potential for early cardiovascular disease (CVD) detection. However, deploying deep learning models for automated analysis in resource-constrained environments faces reliability challenges due to inevitable Out-of-Distribution (OOD) data. OOD inputs, such as unseen pathologies or noisecorrupted signals, often cause erroneous, high-confidence predictions by standard classifiers, compromising patient safety. Existing OOD detection methods either neglect computational constraints or address noise and unseen classes separately. This paper explores Unsupervised Anomaly Detection (UAD) as an independent, upstream filtering mechanism to improve robustness. We benchmark six UAD approaches, including Deep SVDD, reconstruction-based models, Masked Anomaly Detection, normalizing flows, and diffusion models, optimized via Neural Architecture Search (NAS) under strict resource constraints (at most 512k parameters). Evaluation on PTB-XL and BUT QDB datasets assessed detection of OOD CVD classes and signals unsuitable for analysis due to noise. Results show Deep SVDD consistently achieves the best trade-off between detection and efficiency. In a realistic deployment simulation, integrating the optimized Deep SVDD filter with a diagnostic classifier improved accuracy by up to 21 percentage points over a classifier-only baseline. This study demonstrates that optimized UAD filters can safeguard automated ECG analysis, enabling safer, more reliable continuous cardiovascular monitoring on wearables.
Learning ECG Representations via Poly-Window Contrastive Learning
Aug 21, 2025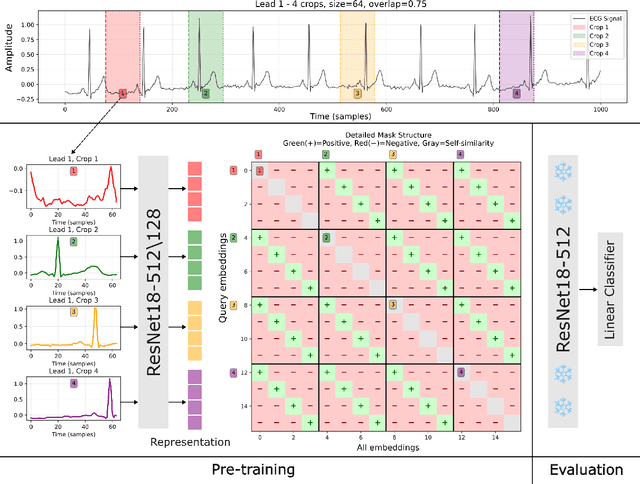
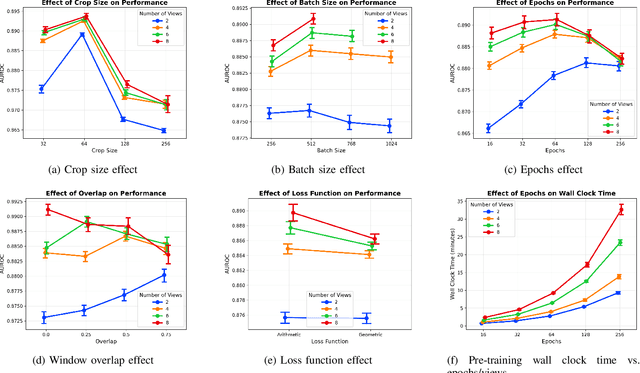
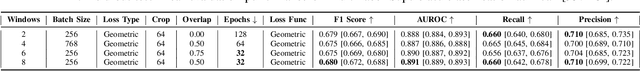

Electrocardiogram (ECG) analysis is foundational for cardiovascular disease diagnosis, yet the performance of deep learning models is often constrained by limited access to annotated data. Self-supervised contrastive learning has emerged as a powerful approach for learning robust ECG representations from unlabeled signals. However, most existing methods generate only pairwise augmented views and fail to leverage the rich temporal structure of ECG recordings. In this work, we present a poly-window contrastive learning framework. We extract multiple temporal windows from each ECG instance to construct positive pairs and maximize their agreement via statistics. Inspired by the principle of slow feature analysis, our approach explicitly encourages the model to learn temporally invariant and physiologically meaningful features that persist across time. We validate our approach through extensive experiments and ablation studies on the PTB-XL dataset. Our results demonstrate that poly-window contrastive learning consistently outperforms conventional two-view methods in multi-label superclass classification, achieving higher AUROC (0.891 vs. 0.888) and F1 scores (0.680 vs. 0.679) while requiring up to four times fewer pre-training epochs (32 vs. 128) and 14.8% in total wall clock pre-training time reduction. Despite processing multiple windows per sample, we achieve a significant reduction in the number of training epochs and total computation time, making our method practical for training foundational models. Through extensive ablations, we identify optimal design choices and demonstrate robustness across various hyperparameters. These findings establish poly-window contrastive learning as a highly efficient and scalable paradigm for automated ECG analysis and provide a promising general framework for self-supervised representation learning in biomedical time-series data.
Bridging Performance Gaps for Foundation Models: A Post-Training Strategy for ECGFounder
Sep 16, 2025ECG foundation models are increasingly popular due to their adaptability across various tasks. However, their clinical applicability is often limited by performance gaps compared to task-specific models, even after pre-training on large ECG datasets and fine-tuning on target data. This limitation is likely due to the lack of an effective post-training strategy. In this paper, we propose a simple yet effective post-training approach to enhance ECGFounder, a state-of-the-art ECG foundation model pre-trained on over 7 million ECG recordings. Experiments on the PTB-XL benchmark show that our approach improves the baseline fine-tuning strategy by 1.2%-3.3% in macro AUROC and 5.3%-20.9% in macro AUPRC. Additionally, our method outperforms several recent state-of-the-art approaches, including task-specific and advanced architectures. Further evaluation reveals that our method is more stable and sample-efficient compared to the baseline, achieving a 9.1% improvement in macro AUROC and a 34.9% improvement in macro AUPRC using just 10% of the training data. Ablation studies identify key components, such as stochastic depth and preview linear probing, that contribute to the enhanced performance. These findings underscore the potential of post-training strategies to improve ECG foundation models, and we hope this work will contribute to the continued development of foundation models in the ECG domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge