Zhiyun Xue
Uncovering the effects of model initialization on deep model generalization: A study with adult and pediatric Chest X-ray images
Sep 20, 2023Abstract:Model initialization techniques are vital for improving the performance and reliability of deep learning models in medical computer vision applications. While much literature exists on non-medical images, the impacts on medical images, particularly chest X-rays (CXRs) are less understood. Addressing this gap, our study explores three deep model initialization techniques: Cold-start, Warm-start, and Shrink and Perturb start, focusing on adult and pediatric populations. We specifically focus on scenarios with periodically arriving data for training, thereby embracing the real-world scenarios of ongoing data influx and the need for model updates. We evaluate these models for generalizability against external adult and pediatric CXR datasets. We also propose novel ensemble methods: F-score-weighted Sequential Least-Squares Quadratic Programming (F-SLSQP) and Attention-Guided Ensembles with Learnable Fuzzy Softmax to aggregate weight parameters from multiple models to capitalize on their collective knowledge and complementary representations. We perform statistical significance tests with 95% confidence intervals and p-values to analyze model performance. Our evaluations indicate models initialized with ImageNet-pre-trained weights demonstrate superior generalizability over randomly initialized counterparts, contradicting some findings for non-medical images. Notably, ImageNet-pretrained models exhibit consistent performance during internal and external testing across different training scenarios. Weight-level ensembles of these models show significantly higher recall (p<0.05) during testing compared to individual models. Thus, our study accentuates the benefits of ImageNet-pretrained weight initialization, especially when used with weight-level ensembles, for creating robust and generalizable deep learning solutions.
Semantically Redundant Training Data Removal and Deep Model Classification Performance: A Study with Chest X-rays
Sep 18, 2023Abstract:Deep learning (DL) has demonstrated its innate capacity to independently learn hierarchical features from complex and multi-dimensional data. A common understanding is that its performance scales up with the amount of training data. Another data attribute is the inherent variety. It follows, therefore, that semantic redundancy, which is the presence of similar or repetitive information, would tend to lower performance and limit generalizability to unseen data. In medical imaging data, semantic redundancy can occur due to the presence of multiple images that have highly similar presentations for the disease of interest. Further, the common use of augmentation methods to generate variety in DL training may be limiting performance when applied to semantically redundant data. We propose an entropy-based sample scoring approach to identify and remove semantically redundant training data. We demonstrate using the publicly available NIH chest X-ray dataset that the model trained on the resulting informative subset of training data significantly outperforms the model trained on the full training set, during both internal (recall: 0.7164 vs 0.6597, p<0.05) and external testing (recall: 0.3185 vs 0.2589, p<0.05). Our findings emphasize the importance of information-oriented training sample selection as opposed to the conventional practice of using all available training data.
Does image resolution impact chest X-ray based fine-grained Tuberculosis-consistent lesion segmentation?
Jan 10, 2023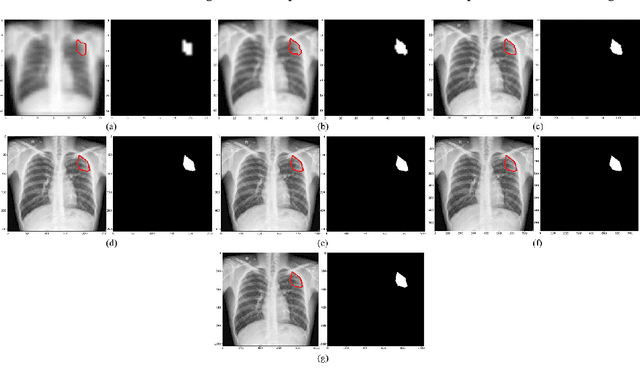
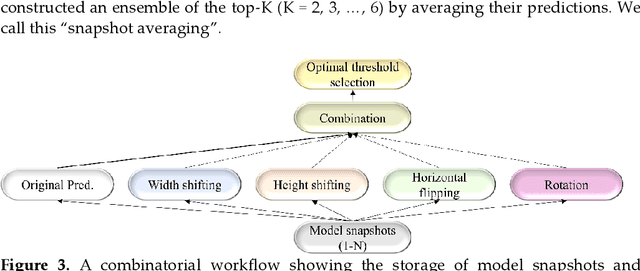
Abstract:Deep learning (DL) models are becoming state-of-the-art in segmenting anatomical and disease regions of interest (ROIs) in medical images, particularly chest X-rays (CXRs). However, these models are reportedly trained on reduced image resolutions citing reasons for the lack of computational resources. Literature is sparse considering identifying the optimal image resolution to train these models for the task under study, particularly considering segmentation of Tuberculosis (TB)-consistent lesions in CXRs. In this study, we used the (i) Shenzhen TB CXR dataset, investigated performance gains achieved through training an Inception-V3-based UNet model using various image/mask resolutions with/without lung ROI cropping and aspect ratio adjustments, and (ii) identified the optimal image resolution through extensive empirical evaluations to improve TB-consistent lesion segmentation performance. We proposed a combinatorial approach consisting of storing model snapshots, optimizing test-time augmentation (TTA) methods, and selecting the optimal segmentation threshold to further improve performance at the optimal resolution. We emphasize that (i) higher image resolutions are not always necessary and (ii) identifying the optimal image resolution is indispensable to achieve superior performance for the task under study.
Generalizability of Deep Adult Lung Segmentation Models to the Pediatric Population: A Retrospective Study
Nov 04, 2022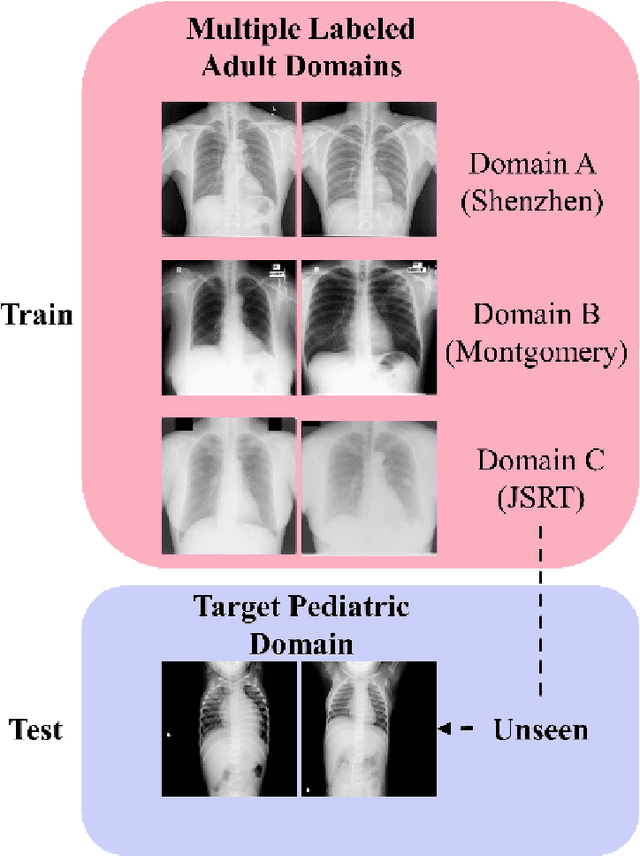


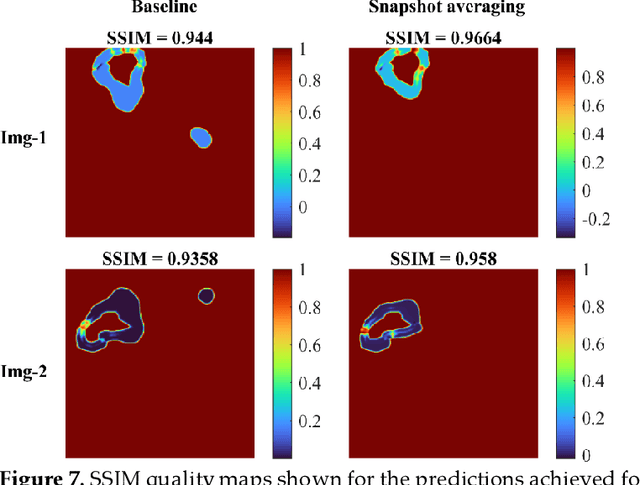
Abstract:Lung segmentation in chest X-rays (CXRs) is an important prerequisite for improving the specificity of diagnoses of cardiopulmonary diseases in a clinical decision support system. Current deep learning (DL) models for lung segmentation are trained and evaluated on CXR datasets in which the radiographic projections are captured predominantly from the adult population. However, the shape of the lungs is reported to be significantly different for pediatrics across the developmental stages from infancy to adulthood. This might result in age-related data domain shifts that would adversely impact lung segmentation performance when the models trained on the adult population are deployed for pediatric lung segmentation. In this work, our goal is to analyze the generalizability of deep adult lung segmentation models to the pediatric population and improve performance through a systematic combinatorial approach consisting of CXR modality-specific weight initializations, stacked generalization, and an ensemble of the stacked generalization models. Novel evaluation metrics consisting of Mean Lung Contour Distance and Average Hash Score are proposed in addition to the Multi-scale Structural Similarity Index Measure, Intersection of Union, and Dice metrics to evaluate segmentation performance. We observed a significant improvement (p < 0.05) in cross-domain generalization through our combinatorial approach. This study could serve as a paradigm to analyze the cross-domain generalizability of deep segmentation models for other medical imaging modalities and applications.
Deep ensemble learning for segmenting tuberculosis-consistent manifestations in chest radiographs
Jun 13, 2022Abstract:Automated segmentation of tuberculosis (TB)-consistent lesions in chest X-rays (CXRs) using deep learning (DL) methods can help reduce radiologist effort, supplement clinical decision-making, and potentially result in improved patient treatment. The majority of works in the literature discuss training automatic segmentation models using coarse bounding box annotations. However, the granularity of the bounding box annotation could result in the inclusion of a considerable fraction of false positives and negatives at the pixel level that may adversely impact overall semantic segmentation performance. This study (i) evaluates the benefits of using fine-grained annotations of TB-consistent lesions and (ii) trains and constructs ensembles of the variants of U-Net models for semantically segmenting TB-consistent lesions in both original and bone-suppressed frontal CXRs. We evaluated segmentation performance using several ensemble methods such as bitwise AND, bitwise-OR, bitwise-MAX, and stacking. We observed that the stacking ensemble demonstrated superior segmentation performance (Dice score: 0.5743, 95% confidence interval: (0.4055,0.7431)) compared to the individual constituent models and other ensemble methods. To the best of our knowledge, this is the first study to apply ensemble learning to improve fine-grained TB-consistent lesion segmentation performance.
Deep Cervix Model Development from Heterogeneous and Partially Labeled Image Datasets
Jan 18, 2022Abstract:Cervical cancer is the fourth most common cancer in women worldwide. The availability of a robust automated cervical image classification system can augment the clinical care provider's limitation in traditional visual inspection with acetic acid (VIA). However, there are a wide variety of cervical inspection objectives which impact the labeling criteria for criteria-specific prediction model development. Moreover, due to the lack of confirmatory test results and inter-rater labeling variation, many images are left unlabeled. Motivated by these challenges, we propose a self-supervised learning (SSL) based approach to produce a pre-trained cervix model from unlabeled cervical images. The developed model is further fine-tuned to produce criteria-specific classification models with the available labeled images. We demonstrate the effectiveness of the proposed approach using two cervical image datasets. Both datasets are partially labeled and labeling criteria are different. The experimental results show that the SSL-based initialization improves classification performance (Accuracy: 2.5% min) and the inclusion of images from both datasets during SSL further improves the performance (Accuracy: 1.5% min). Further, considering data-sharing restrictions, we experimented with the effectiveness of Federated SSL and find that it can improve performance over the SSL model developed with just its images. This justifies the importance of SSL-based cervix model development. We believe that the present research shows a novel direction in developing criteria-specific custom deep models for cervical image classification by combining images from different sources unlabeled and/or labeled with varying criteria, and addressing image access restrictions.
Selective Synthetic Augmentation with HistoGAN for Improved Histopathology Image Classification
Nov 10, 2021



Abstract:Histopathological analysis is the present gold standard for precancerous lesion diagnosis. The goal of automated histopathological classification from digital images requires supervised training, which requires a large number of expert annotations that can be expensive and time-consuming to collect. Meanwhile, accurate classification of image patches cropped from whole-slide images is essential for standard sliding window based histopathology slide classification methods. To mitigate these issues, we propose a carefully designed conditional GAN model, namely HistoGAN, for synthesizing realistic histopathology image patches conditioned on class labels. We also investigate a novel synthetic augmentation framework that selectively adds new synthetic image patches generated by our proposed HistoGAN, rather than expanding directly the training set with synthetic images. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation. Our models are evaluated on two datasets: a cervical histopathology image dataset with limited annotations, and another dataset of lymph node histopathology images with metastatic cancer. Here, we show that leveraging HistoGAN generated images with selective augmentation results in significant and consistent improvements of classification performance (6.7% and 2.8% higher accuracy, respectively) for cervical histopathology and metastatic cancer datasets.
* Elsevier Medical Image Analysis Best Paper Award runner up. arXiv admin note: substantial text overlap with arXiv:1912.03837
Synthetic Sample Selection via Reinforcement Learning
Aug 26, 2020



Abstract:Synthesizing realistic medical images provides a feasible solution to the shortage of training data in deep learning based medical image recognition systems. However, the quality control of synthetic images for data augmentation purposes is under-investigated, and some of the generated images are not realistic and may contain misleading features that distort data distribution when mixed with real images. Thus, the effectiveness of those synthetic images in medical image recognition systems cannot be guaranteed when they are being added randomly without quality assurance. In this work, we propose a reinforcement learning (RL) based synthetic sample selection method that learns to choose synthetic images containing reliable and informative features. A transformer based controller is trained via proximal policy optimization (PPO) using the validation classification accuracy as the reward. The selected images are mixed with the original training data for improved training of image recognition systems. To validate our method, we take the pathology image recognition as an example and conduct extensive experiments on two histopathology image datasets. In experiments on a cervical dataset and a lymph node dataset, the image classification performance is improved by 8.1% and 2.3%, respectively, when utilizing high-quality synthetic images selected by our RL framework. Our proposed synthetic sample selection method is general and has great potential to boost the performance of various medical image recognition systems given limited annotation.
Feature based Sequential Classifier with Attention Mechanism
Jul 22, 2020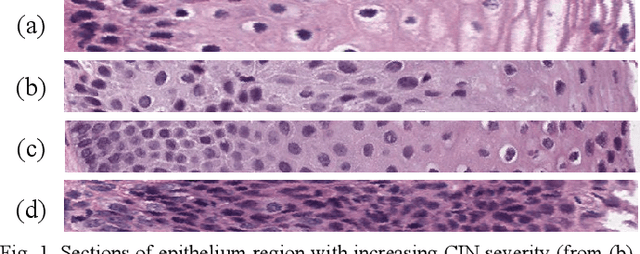
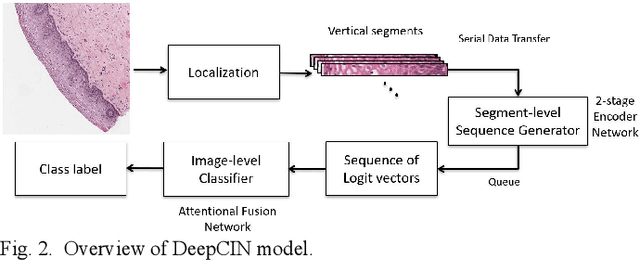
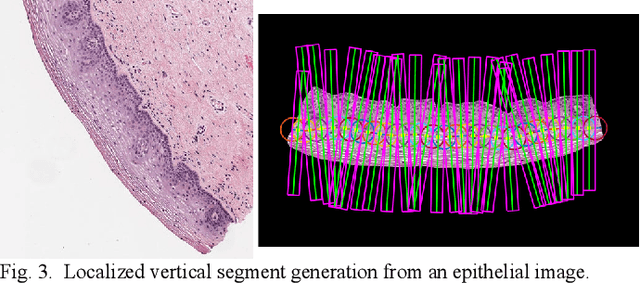
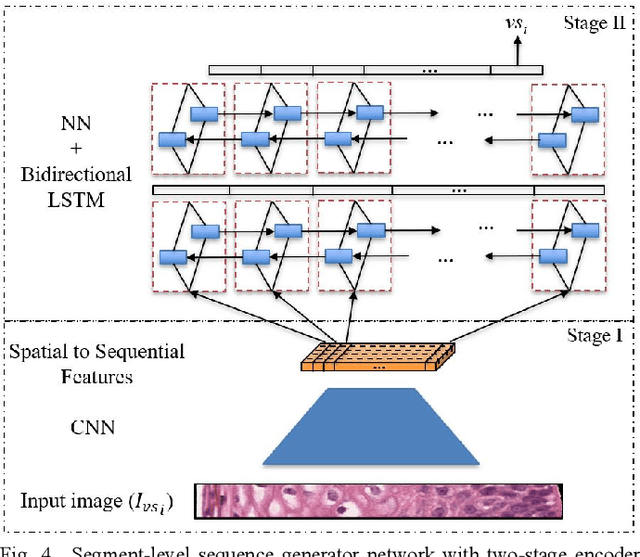
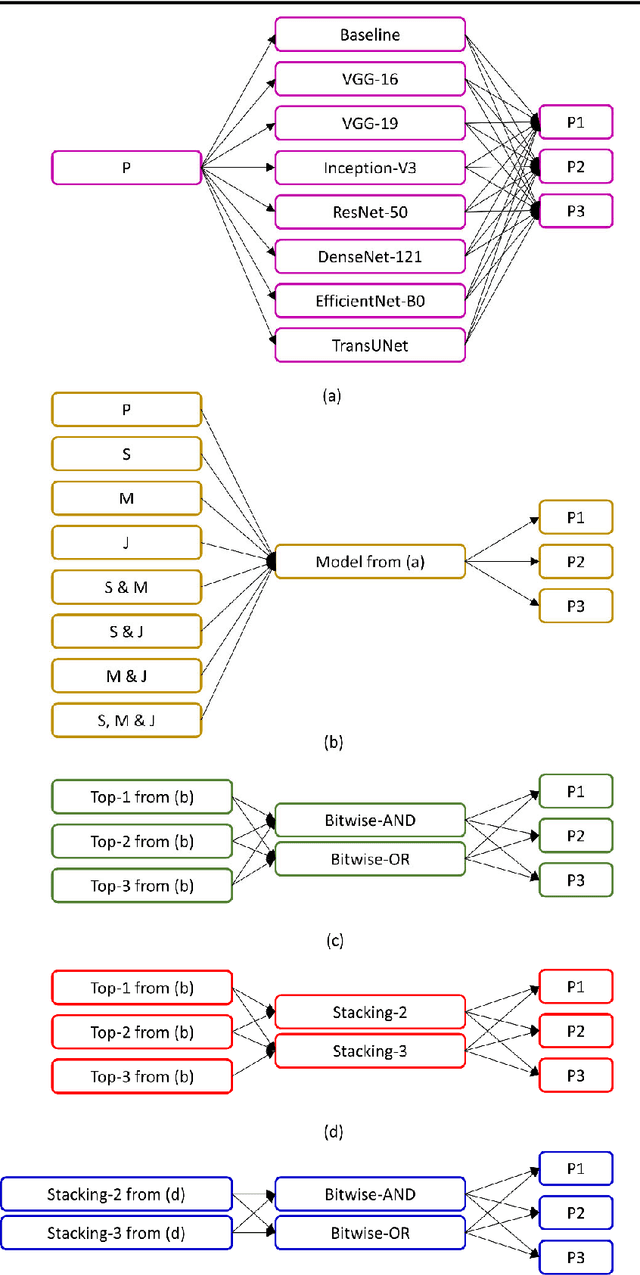
Abstract:Cervical cancer is one of the deadliest cancers affecting women globally. Cervical intraepithelial neoplasia (CIN) assessment using histopathological examination of cervical biopsy slides is subject to interobserver variability. Automated processing of digitized histopathology slides has the potential for more accurate classification for CIN grades from normal to increasing grades of pre-malignancy: CIN1, CIN2 and CIN3. Cervix disease is generally understood to progress from the bottom (basement membrane) to the top of the epithelium. To model this relationship of disease severity to spatial distribution of abnormalities, we propose a network pipeline, DeepCIN, to analyze high-resolution epithelium images (manually extracted from whole-slide images) hierarchically by focusing on localized vertical regions and fusing this local information for determining Normal/CIN classification. The pipeline contains two classifier networks: 1) a cross-sectional, vertical segment-level sequence generator (two-stage encoder model) is trained using weak supervision to generate feature sequences from the vertical segments to preserve the bottom-to-top feature relationships in the epithelium image data; 2) an attention-based fusion network image-level classifier predicting the final CIN grade by merging vertical segment sequences. The model produces the CIN classification results and also determines the vertical segment contributions to CIN grade prediction. Experiments show that DeepCIN achieves pathologist-level CIN classification accuracy.
Selective Synthetic Augmentation with Quality Assurance
Dec 09, 2019



Abstract:Supervised training of an automated medical image analysis system often requires a large amount of expert annotations that are hard to collect. Moreover, the proportions of data available across different classes may be highly imbalanced for rare diseases. To mitigate these issues, we investigate a novel data augmentation pipeline that selectively adds new synthetic images generated by conditional Adversarial Networks (cGANs), rather than extending directly the training set with synthetic images. The selection mechanisms that we introduce to the synthetic augmentation pipeline are motivated by the observation that, although cGAN-generated images can be visually appealing, they are not guaranteed to contain essential features for classification performance improvement. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation by ensuring that adding the selected synthetic images to the training set will improve performance. We evaluate our model on a medical histopathology dataset, and two natural image classification benchmarks, CIFAR10 and SVHN. Results on these datasets show significant and consistent improvements in classification performance (with 6.8%, 3.9%, 1.6% higher accuracy, respectively) by leveraging cGAN generated images with selective augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge