Sudhir Sornapudi
Corteva Agriscience, Indianapolis, USA
Few-Shot Adaptation of Grounding DINO for Agricultural Domain
Apr 09, 2025Abstract:Deep learning models are transforming agricultural applications by enabling automated phenotyping, monitoring, and yield estimation. However, their effectiveness heavily depends on large amounts of annotated training data, which can be labor and time intensive. Recent advances in open-set object detection, particularly with models like Grounding-DINO, offer a potential solution to detect regions of interests based on text prompt input. Initial zero-shot experiments revealed challenges in crafting effective text prompts, especially for complex objects like individual leaves and visually similar classes. To address these limitations, we propose an efficient few-shot adaptation method that simplifies the Grounding-DINO architecture by removing the text encoder module (BERT) and introducing a randomly initialized trainable text embedding. This method achieves superior performance across multiple agricultural datasets, including plant-weed detection, plant counting, insect identification, fruit counting, and remote sensing tasks. Specifically, it demonstrates up to a $\sim24\%$ higher mAP than fully fine-tuned YOLO models on agricultural datasets and outperforms previous state-of-the-art methods by $\sim10\%$ in remote sensing, under few-shot learning conditions. Our method offers a promising solution for automating annotation and accelerating the development of specialized agricultural AI solutions.
Self-Supervised Backbone Framework for Diverse Agricultural Vision Tasks
Mar 22, 2024



Abstract:Computer vision in agriculture is game-changing with its ability to transform farming into a data-driven, precise, and sustainable industry. Deep learning has empowered agriculture vision to analyze vast, complex visual data, but heavily rely on the availability of large annotated datasets. This remains a bottleneck as manual labeling is error-prone, time-consuming, and expensive. The lack of efficient labeling approaches inspired us to consider self-supervised learning as a paradigm shift, learning meaningful feature representations from raw agricultural image data. In this work, we explore how self-supervised representation learning unlocks the potential applicability to diverse agriculture vision tasks by eliminating the need for large-scale annotated datasets. We propose a lightweight framework utilizing SimCLR, a contrastive learning approach, to pre-train a ResNet-50 backbone on a large, unannotated dataset of real-world agriculture field images. Our experimental analysis and results indicate that the model learns robust features applicable to a broad range of downstream agriculture tasks discussed in the paper. Additionally, the reduced reliance on annotated data makes our approach more cost-effective and accessible, paving the way for broader adoption of computer vision in agriculture.
Feature based Sequential Classifier with Attention Mechanism
Jul 22, 2020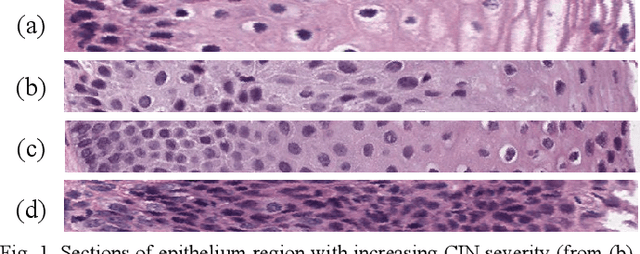
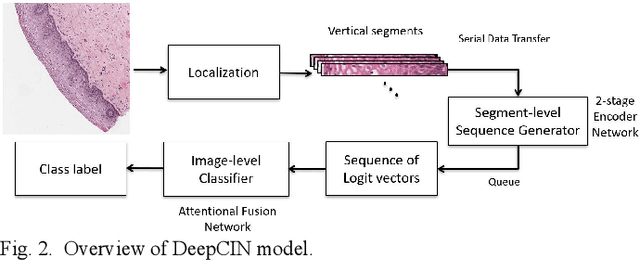
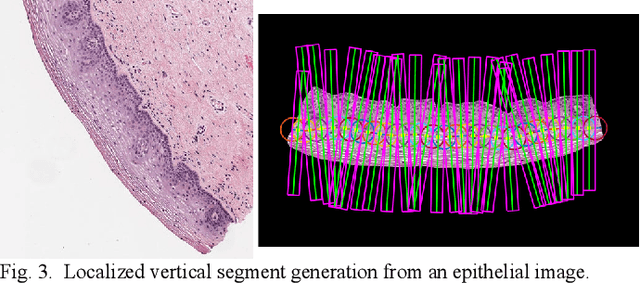
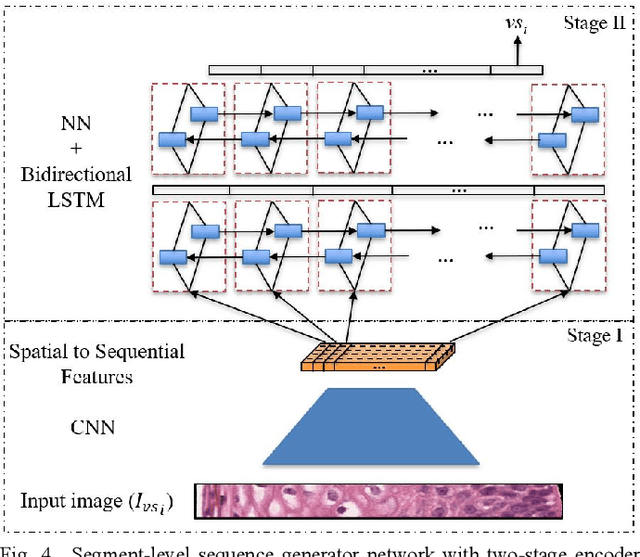
Abstract:Cervical cancer is one of the deadliest cancers affecting women globally. Cervical intraepithelial neoplasia (CIN) assessment using histopathological examination of cervical biopsy slides is subject to interobserver variability. Automated processing of digitized histopathology slides has the potential for more accurate classification for CIN grades from normal to increasing grades of pre-malignancy: CIN1, CIN2 and CIN3. Cervix disease is generally understood to progress from the bottom (basement membrane) to the top of the epithelium. To model this relationship of disease severity to spatial distribution of abnormalities, we propose a network pipeline, DeepCIN, to analyze high-resolution epithelium images (manually extracted from whole-slide images) hierarchically by focusing on localized vertical regions and fusing this local information for determining Normal/CIN classification. The pipeline contains two classifier networks: 1) a cross-sectional, vertical segment-level sequence generator (two-stage encoder model) is trained using weak supervision to generate feature sequences from the vertical segments to preserve the bottom-to-top feature relationships in the epithelium image data; 2) an attention-based fusion network image-level classifier predicting the final CIN grade by merging vertical segment sequences. The model produces the CIN classification results and also determines the vertical segment contributions to CIN grade prediction. Experiments show that DeepCIN achieves pathologist-level CIN classification accuracy.
Comparing Deep Learning Models for Multi-cell Classification in Liquid-based Cervical Cytology Images
Oct 02, 2019



Abstract:Liquid-based cytology (LBC) is a reliable automated technique for the screening of Papanicolaou (Pap) smear data. It is an effective technique for collecting a majority of the cervical cells and aiding cytopathologists in locating abnormal cells. Most methods published in the research literature rely on accurate cell segmentation as a prior, which remains challenging due to a variety of factors, e.g., stain consistency, presence of clustered cells, etc. We propose a method for automatic classification of cervical slide images through generation of labeled cervical patch data and extracting deep hierarchical features by fine-tuning convolution neural networks, as well as a novel graph-based cell detection approach for cellular level evaluation. The results show that the proposed pipeline can classify images of both single cell and overlapping cells. The VGG-19 model is found to be the best at classifying the cervical cytology patch data with 95 % accuracy under precision-recall curve.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge