Rodney Long
Functional Parcellation of fMRI data using multistage k-means clustering
Feb 19, 2022Abstract:Purpose: Functional Magnetic Resonance Imaging (fMRI) data acquired through resting-state studies have been used to obtain information about the spontaneous activations inside the brain. One of the approaches for analysis and interpretation of resting-state fMRI data require spatially and functionally homogenous parcellation of the whole brain based on underlying temporal fluctuations. Clustering is often used to generate functional parcellation. However, major clustering algorithms, when used for fMRI data, have their limitations. Among commonly used parcellation schemes, a tradeoff exists between intra-cluster functional similarity and alignment with anatomical regions. Approach: In this work, we present a clustering algorithm for resting state and task fMRI data which is developed to obtain brain parcellations that show high structural and functional homogeneity. The clustering is performed by multistage binary k-means clustering algorithm designed specifically for the 4D fMRI data. The results from this multistage k-means algorithm show that by modifying and combining different algorithms, we can take advantage of the strengths of different techniques while overcoming their limitations. Results: The clustering output for resting state fMRI data using the multistage k-means approach is shown to be better than simple k-means or functional atlas in terms of spatial and functional homogeneity. The clusters also correspond to commonly identifiable brain networks. For task fMRI, the clustering output can identify primary and secondary activation regions and provide information about the varying hemodynamic response across different brain regions. Conclusion: The multistage k-means approach can provide functional parcellations of the brain using resting state fMRI data. The method is model-free and is data driven which can be applied to both resting state and task fMRI.
Selective Synthetic Augmentation with HistoGAN for Improved Histopathology Image Classification
Nov 10, 2021



Abstract:Histopathological analysis is the present gold standard for precancerous lesion diagnosis. The goal of automated histopathological classification from digital images requires supervised training, which requires a large number of expert annotations that can be expensive and time-consuming to collect. Meanwhile, accurate classification of image patches cropped from whole-slide images is essential for standard sliding window based histopathology slide classification methods. To mitigate these issues, we propose a carefully designed conditional GAN model, namely HistoGAN, for synthesizing realistic histopathology image patches conditioned on class labels. We also investigate a novel synthetic augmentation framework that selectively adds new synthetic image patches generated by our proposed HistoGAN, rather than expanding directly the training set with synthetic images. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation. Our models are evaluated on two datasets: a cervical histopathology image dataset with limited annotations, and another dataset of lymph node histopathology images with metastatic cancer. Here, we show that leveraging HistoGAN generated images with selective augmentation results in significant and consistent improvements of classification performance (6.7% and 2.8% higher accuracy, respectively) for cervical histopathology and metastatic cancer datasets.
* Elsevier Medical Image Analysis Best Paper Award runner up. arXiv admin note: substantial text overlap with arXiv:1912.03837
Feature based Sequential Classifier with Attention Mechanism
Jul 22, 2020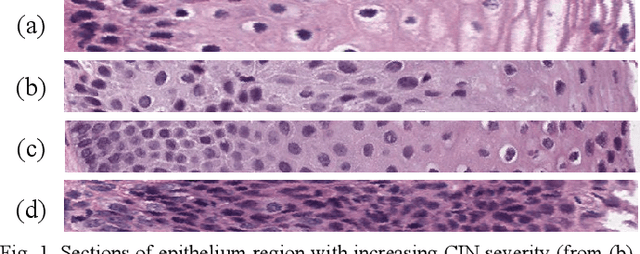
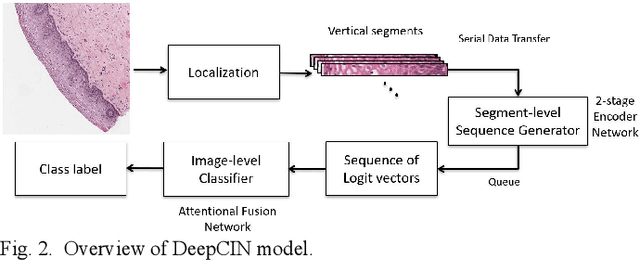
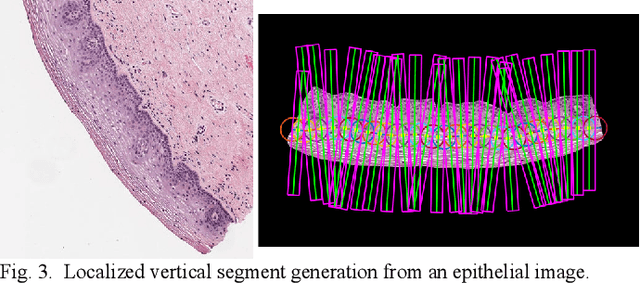
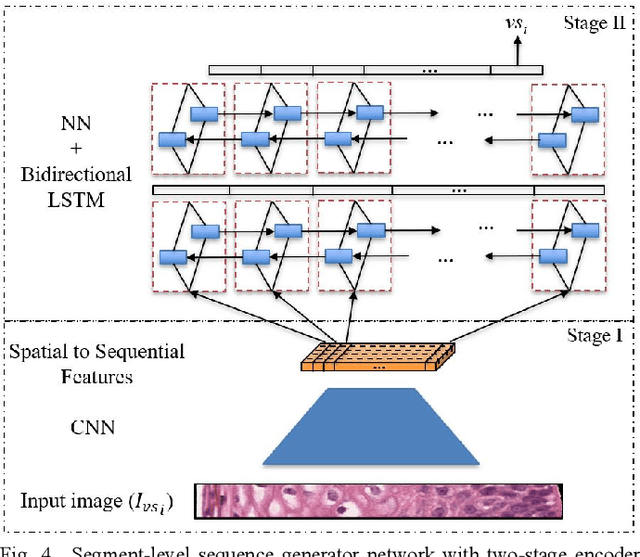
Abstract:Cervical cancer is one of the deadliest cancers affecting women globally. Cervical intraepithelial neoplasia (CIN) assessment using histopathological examination of cervical biopsy slides is subject to interobserver variability. Automated processing of digitized histopathology slides has the potential for more accurate classification for CIN grades from normal to increasing grades of pre-malignancy: CIN1, CIN2 and CIN3. Cervix disease is generally understood to progress from the bottom (basement membrane) to the top of the epithelium. To model this relationship of disease severity to spatial distribution of abnormalities, we propose a network pipeline, DeepCIN, to analyze high-resolution epithelium images (manually extracted from whole-slide images) hierarchically by focusing on localized vertical regions and fusing this local information for determining Normal/CIN classification. The pipeline contains two classifier networks: 1) a cross-sectional, vertical segment-level sequence generator (two-stage encoder model) is trained using weak supervision to generate feature sequences from the vertical segments to preserve the bottom-to-top feature relationships in the epithelium image data; 2) an attention-based fusion network image-level classifier predicting the final CIN grade by merging vertical segment sequences. The model produces the CIN classification results and also determines the vertical segment contributions to CIN grade prediction. Experiments show that DeepCIN achieves pathologist-level CIN classification accuracy.
Selective Synthetic Augmentation with Quality Assurance
Dec 09, 2019



Abstract:Supervised training of an automated medical image analysis system often requires a large amount of expert annotations that are hard to collect. Moreover, the proportions of data available across different classes may be highly imbalanced for rare diseases. To mitigate these issues, we investigate a novel data augmentation pipeline that selectively adds new synthetic images generated by conditional Adversarial Networks (cGANs), rather than extending directly the training set with synthetic images. The selection mechanisms that we introduce to the synthetic augmentation pipeline are motivated by the observation that, although cGAN-generated images can be visually appealing, they are not guaranteed to contain essential features for classification performance improvement. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation by ensuring that adding the selected synthetic images to the training set will improve performance. We evaluate our model on a medical histopathology dataset, and two natural image classification benchmarks, CIFAR10 and SVHN. Results on these datasets show significant and consistent improvements in classification performance (with 6.8%, 3.9%, 1.6% higher accuracy, respectively) by leveraging cGAN generated images with selective augmentation.
Comparing Deep Learning Models for Multi-cell Classification in Liquid-based Cervical Cytology Images
Oct 02, 2019



Abstract:Liquid-based cytology (LBC) is a reliable automated technique for the screening of Papanicolaou (Pap) smear data. It is an effective technique for collecting a majority of the cervical cells and aiding cytopathologists in locating abnormal cells. Most methods published in the research literature rely on accurate cell segmentation as a prior, which remains challenging due to a variety of factors, e.g., stain consistency, presence of clustered cells, etc. We propose a method for automatic classification of cervical slide images through generation of labeled cervical patch data and extracting deep hierarchical features by fine-tuning convolution neural networks, as well as a novel graph-based cell detection approach for cellular level evaluation. The results show that the proposed pipeline can classify images of both single cell and overlapping cells. The VGG-19 model is found to be the best at classifying the cervical cytology patch data with 95 % accuracy under precision-recall curve.
SegAN: Adversarial Network with Multi-scale $L_1$ Loss for Medical Image Segmentation
Jul 16, 2017



Abstract:Inspired by classic generative adversarial networks (GAN), we propose a novel end-to-end adversarial neural network, called SegAN, for the task of medical image segmentation. Since image segmentation requires dense, pixel-level labeling, the single scalar real/fake output of a classic GAN's discriminator may be ineffective in producing stable and sufficient gradient feedback to the networks. Instead, we use a fully convolutional neural network as the segmentor to generate segmentation label maps, and propose a novel adversarial critic network with a multi-scale $L_1$ loss function to force the critic and segmentor to learn both global and local features that capture long- and short-range spatial relationships between pixels. In our SegAN framework, the segmentor and critic networks are trained in an alternating fashion in a min-max game: The critic takes as input a pair of images, (original_image $*$ predicted_label_map, original_image $*$ ground_truth_label_map), and then is trained by maximizing a multi-scale loss function; The segmentor is trained with only gradients passed along by the critic, with the aim to minimize the multi-scale loss function. We show that such a SegAN framework is more effective and stable for the segmentation task, and it leads to better performance than the state-of-the-art U-net segmentation method. We tested our SegAN method using datasets from the MICCAI BRATS brain tumor segmentation challenge. Extensive experimental results demonstrate the effectiveness of the proposed SegAN with multi-scale loss: on BRATS 2013 SegAN gives performance comparable to the state-of-the-art for whole tumor and tumor core segmentation while achieves better precision and sensitivity for Gd-enhance tumor core segmentation; on BRATS 2015 SegAN achieves better performance than the state-of-the-art in both dice score and precision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge