Jiarong Ye
Synthetic Augmentation with Large-scale Unconditional Pre-training
Aug 08, 2023



Abstract:Deep learning based medical image recognition systems often require a substantial amount of training data with expert annotations, which can be expensive and time-consuming to obtain. Recently, synthetic augmentation techniques have been proposed to mitigate the issue by generating realistic images conditioned on class labels. However, the effectiveness of these methods heavily depends on the representation capability of the trained generative model, which cannot be guaranteed without sufficient labeled training data. To further reduce the dependency on annotated data, we propose a synthetic augmentation method called HistoDiffusion, which can be pre-trained on large-scale unlabeled datasets and later applied to a small-scale labeled dataset for augmented training. In particular, we train a latent diffusion model (LDM) on diverse unlabeled datasets to learn common features and generate realistic images without conditional inputs. Then, we fine-tune the model with classifier guidance in latent space on an unseen labeled dataset so that the model can synthesize images of specific categories. Additionally, we adopt a selective mechanism to only add synthetic samples with high confidence of matching to target labels. We evaluate our proposed method by pre-training on three histopathology datasets and testing on a histopathology dataset of colorectal cancer (CRC) excluded from the pre-training datasets. With HistoDiffusion augmentation, the classification accuracy of a backbone classifier is remarkably improved by 6.4% using a small set of the original labels. Our code is available at https://github.com/karenyyy/HistoDiffAug.
Accurate Virus Identification with Interpretable Raman Signatures by Machine Learning
Jun 05, 2022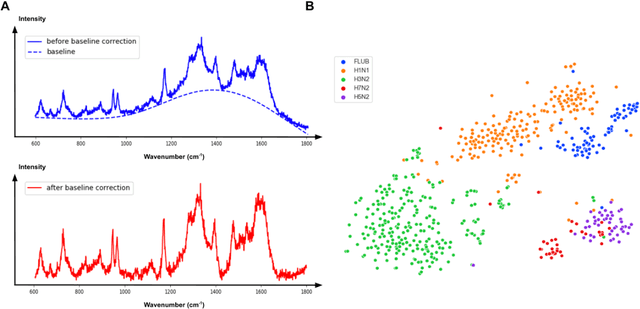
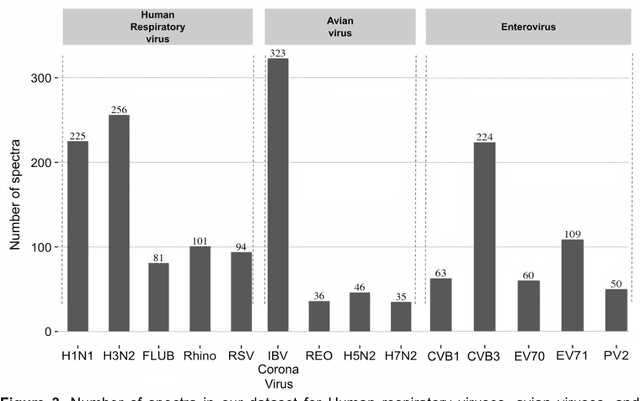
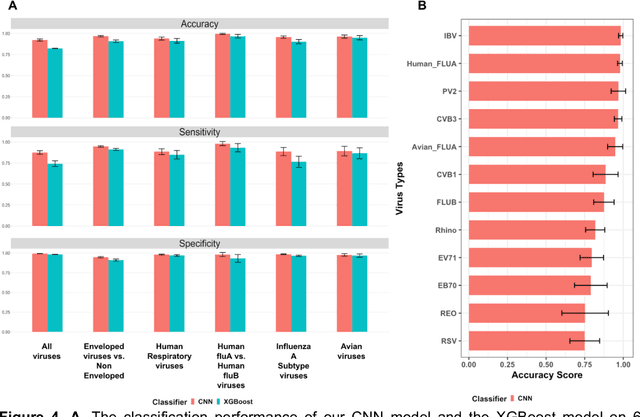
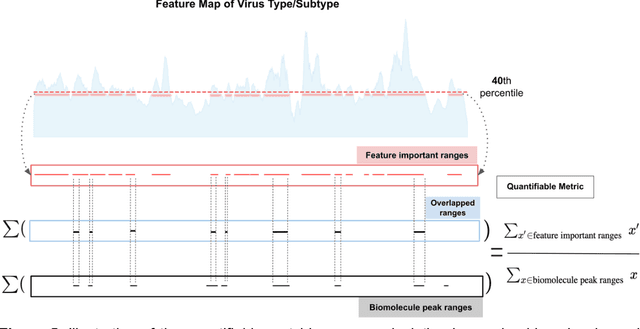
Abstract:Rapid identification of newly emerging or circulating viruses is an important first step toward managing the public health response to potential outbreaks. A portable virus capture device coupled with label-free Raman Spectroscopy holds the promise of fast detection by rapidly obtaining the Raman signature of a virus followed by a machine learning approach applied to recognize the virus based on its Raman spectrum, which is used as a fingerprint. We present such a machine learning approach for analyzing Raman spectra of human and avian viruses. A Convolutional Neural Network (CNN) classifier specifically designed for spectral data achieves very high accuracy for a variety of virus type or subtype identification tasks. In particular, it achieves 99% accuracy for classifying influenza virus type A vs. type B, 96% accuracy for classifying four subtypes of influenza A, 95% accuracy for differentiating enveloped and non-enveloped viruses, and 99% accuracy for differentiating avian coronavirus (infectious bronchitis virus, IBV) from other avian viruses. Furthermore, interpretation of neural net responses in the trained CNN model using a full-gradient algorithm highlights Raman spectral ranges that are most important to virus identification. By correlating ML-selected salient Raman ranges with the signature ranges of known biomolecules and chemical functional groups (for example, amide, amino acid, carboxylic acid), we verify that our ML model effectively recognizes the Raman signatures of proteins, lipids and other vital functional groups present in different viruses and uses a weighted combination of these signatures to identify viruses.
* 23 pages, 8 figures
Selective Synthetic Augmentation with HistoGAN for Improved Histopathology Image Classification
Nov 10, 2021



Abstract:Histopathological analysis is the present gold standard for precancerous lesion diagnosis. The goal of automated histopathological classification from digital images requires supervised training, which requires a large number of expert annotations that can be expensive and time-consuming to collect. Meanwhile, accurate classification of image patches cropped from whole-slide images is essential for standard sliding window based histopathology slide classification methods. To mitigate these issues, we propose a carefully designed conditional GAN model, namely HistoGAN, for synthesizing realistic histopathology image patches conditioned on class labels. We also investigate a novel synthetic augmentation framework that selectively adds new synthetic image patches generated by our proposed HistoGAN, rather than expanding directly the training set with synthetic images. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation. Our models are evaluated on two datasets: a cervical histopathology image dataset with limited annotations, and another dataset of lymph node histopathology images with metastatic cancer. Here, we show that leveraging HistoGAN generated images with selective augmentation results in significant and consistent improvements of classification performance (6.7% and 2.8% higher accuracy, respectively) for cervical histopathology and metastatic cancer datasets.
* Elsevier Medical Image Analysis Best Paper Award runner up. arXiv admin note: substantial text overlap with arXiv:1912.03837
A Multi-attribute Controllable Generative Model for Histopathology Image Synthesis
Nov 10, 2021



Abstract:Generative models have been applied in the medical imaging domain for various image recognition and synthesis tasks. However, a more controllable and interpretable image synthesis model is still lacking yet necessary for important applications such as assisting in medical training. In this work, we leverage the efficient self-attention and contrastive learning modules and build upon state-of-the-art generative adversarial networks (GANs) to achieve an attribute-aware image synthesis model, termed AttributeGAN, which can generate high-quality histopathology images based on multi-attribute inputs. In comparison to existing single-attribute conditional generative models, our proposed model better reflects input attributes and enables smoother interpolation among attribute values. We conduct experiments on a histopathology dataset containing stained H&E images of urothelial carcinoma and demonstrate the effectiveness of our proposed model via comprehensive quantitative and qualitative comparisons with state-of-the-art models as well as different variants of our model. Code is available at https://github.com/karenyyy/MICCAI2021AttributeGAN.
Synthetic Sample Selection via Reinforcement Learning
Aug 26, 2020



Abstract:Synthesizing realistic medical images provides a feasible solution to the shortage of training data in deep learning based medical image recognition systems. However, the quality control of synthetic images for data augmentation purposes is under-investigated, and some of the generated images are not realistic and may contain misleading features that distort data distribution when mixed with real images. Thus, the effectiveness of those synthetic images in medical image recognition systems cannot be guaranteed when they are being added randomly without quality assurance. In this work, we propose a reinforcement learning (RL) based synthetic sample selection method that learns to choose synthetic images containing reliable and informative features. A transformer based controller is trained via proximal policy optimization (PPO) using the validation classification accuracy as the reward. The selected images are mixed with the original training data for improved training of image recognition systems. To validate our method, we take the pathology image recognition as an example and conduct extensive experiments on two histopathology image datasets. In experiments on a cervical dataset and a lymph node dataset, the image classification performance is improved by 8.1% and 2.3%, respectively, when utilizing high-quality synthetic images selected by our RL framework. Our proposed synthetic sample selection method is general and has great potential to boost the performance of various medical image recognition systems given limited annotation.
Selective Synthetic Augmentation with Quality Assurance
Dec 09, 2019



Abstract:Supervised training of an automated medical image analysis system often requires a large amount of expert annotations that are hard to collect. Moreover, the proportions of data available across different classes may be highly imbalanced for rare diseases. To mitigate these issues, we investigate a novel data augmentation pipeline that selectively adds new synthetic images generated by conditional Adversarial Networks (cGANs), rather than extending directly the training set with synthetic images. The selection mechanisms that we introduce to the synthetic augmentation pipeline are motivated by the observation that, although cGAN-generated images can be visually appealing, they are not guaranteed to contain essential features for classification performance improvement. By selecting synthetic images based on the confidence of their assigned labels and their feature similarity to real labeled images, our framework provides quality assurance to synthetic augmentation by ensuring that adding the selected synthetic images to the training set will improve performance. We evaluate our model on a medical histopathology dataset, and two natural image classification benchmarks, CIFAR10 and SVHN. Results on these datasets show significant and consistent improvements in classification performance (with 6.8%, 3.9%, 1.6% higher accuracy, respectively) by leveraging cGAN generated images with selective augmentation.
Synthetic Augmentation and Feature-based Filtering for Improved Cervical Histopathology Image Classification
Jul 24, 2019



Abstract:Cervical intraepithelial neoplasia (CIN) grade of histopathology images is a crucial indicator in cervical biopsy results. Accurate CIN grading of epithelium regions helps pathologists with precancerous lesion diagnosis and treatment planning. Although an automated CIN grading system has been desired, supervised training of such a system would require a large amount of expert annotations, which are expensive and time-consuming to collect. In this paper, we investigate the CIN grade classification problem on segmented epithelium patches. We propose to use conditional Generative Adversarial Networks (cGANs) to expand the limited training dataset, by synthesizing realistic cervical histopathology images. While the synthetic images are visually appealing, they are not guaranteed to contain meaningful features for data augmentation. To tackle this issue, we propose a synthetic-image filtering mechanism based on the divergence in feature space between generated images and class centroids in order to control the feature quality of selected synthetic images for data augmentation. Our models are evaluated on a cervical histopathology image dataset with a limited number of patch-level CIN grade annotations. Extensive experimental results show a significant improvement of classification accuracy from 66.3% to 71.7% using the same ResNet18 baseline classifier after leveraging our cGAN generated images with feature-based filtering, which demonstrates the effectiveness of our models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge