Zhuohang Yu
Inorganic Catalyst Efficiency Prediction Based on EAPCR Model: A Deep Learning Solution for Multi-Source Heterogeneous Data
Mar 10, 2025



Abstract:The design of inorganic catalysts and the prediction of their catalytic efficiency are fundamental challenges in chemistry and materials science. Traditional catalyst evaluation methods primarily rely on machine learning techniques; however, these methods often struggle to process multi-source heterogeneous data, limiting both predictive accuracy and generalization. To address these limitations, this study introduces the Embedding-Attention-Permutated CNN-Residual (EAPCR) deep learning model. EAPCR constructs a feature association matrix using embedding and attention mechanisms and enhances predictive performance through permutated CNN architectures and residual connections. This approach enables the model to accurately capture complex feature interactions across various catalytic conditions, leading to precise efficiency predictions. EAPCR serves as a powerful tool for computational researchers while also assisting domain experts in optimizing catalyst design, effectively bridging the gap between data-driven modeling and experimental applications. We evaluate EAPCR on datasets from TiO2 photocatalysis, thermal catalysis, and electrocatalysis, demonstrating its superiority over traditional machine learning methods (e.g., linear regression, random forest) as well as conventional deep learning models (e.g., ANN, NNs). Across multiple evaluation metrics (MAE, MSE, R2, and RMSE), EAPCR consistently outperforms existing approaches. These findings highlight the strong potential of EAPCR in inorganic catalytic efficiency prediction. As a versatile deep learning framework, EAPCR not only improves predictive accuracy but also establishes a solid foundation for future large-scale model development in inorganic catalysis.
Matching Free Depth Recovery from Structured Light
Jan 13, 2025



Abstract:We present a novel approach for depth estimation from images captured by structured light systems. Unlike many previous methods that rely on image matching process, our approach uses a density voxel grid to represent scene geometry, which is trained via self-supervised differentiable volume rendering. Our method leverages color fields derived from projected patterns in structured light systems during the rendering process, enabling the isolated optimization of the geometry field. This contributes to faster convergence and high-quality output. Additionally, we incorporate normalized device coordinates (NDC), a distortion loss, and a novel surface-based color loss to enhance geometric fidelity. Experimental results demonstrate that our method outperforms existing matching-based techniques in geometric performance for few-shot scenarios, achieving approximately a 60% reduction in average estimated depth errors on synthetic scenes and about 30% on real-world captured scenes. Furthermore, our approach delivers fast training, with a speed roughly three times faster than previous matching-free methods that employ implicit representations.
EAPCR: A Universal Feature Extractor for Scientific Data without Explicit Feature Relation Patterns
Nov 12, 2024



Abstract:Conventional methods, including Decision Tree (DT)-based methods, have been effective in scientific tasks, such as non-image medical diagnostics, system anomaly detection, and inorganic catalysis efficiency prediction. However, most deep-learning techniques have struggled to surpass or even match this level of success as traditional machine-learning methods. The primary reason is that these applications involve multi-source, heterogeneous data where features lack explicit relationships. This contrasts with image data, where pixels exhibit spatial relationships; textual data, where words have sequential dependencies; and graph data, where nodes are connected through established associations. The absence of explicit Feature Relation Patterns (FRPs) presents a significant challenge for deep learning techniques in scientific applications that are not image, text, and graph-based. In this paper, we introduce EAPCR, a universal feature extractor designed for data without explicit FRPs. Tested across various scientific tasks, EAPCR consistently outperforms traditional methods and bridges the gap where deep learning models fall short. To further demonstrate its robustness, we synthesize a dataset without explicit FRPs. While Kolmogorov-Arnold Network (KAN) and feature extractors like Convolutional Neural Networks (CNNs), Graph Convolutional Networks (GCNs), and Transformers struggle, EAPCR excels, demonstrating its robustness and superior performance in scientific tasks without FRPs.
Accurate Virus Identification with Interpretable Raman Signatures by Machine Learning
Jun 05, 2022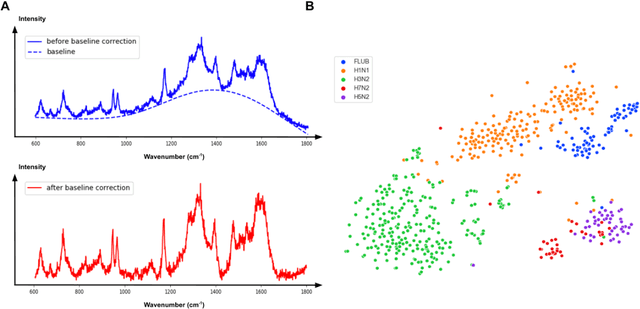
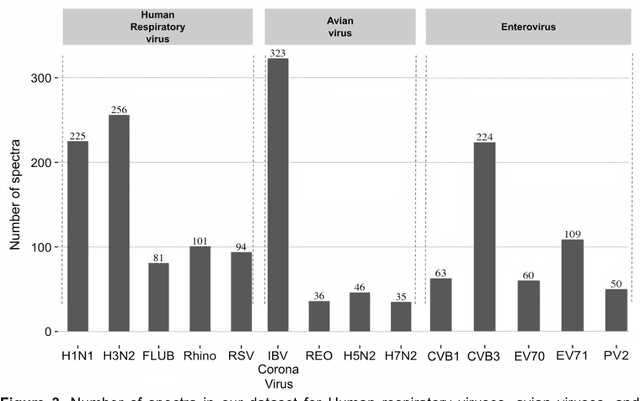
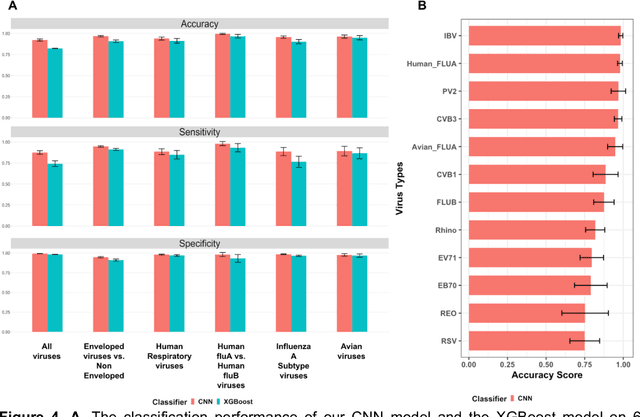
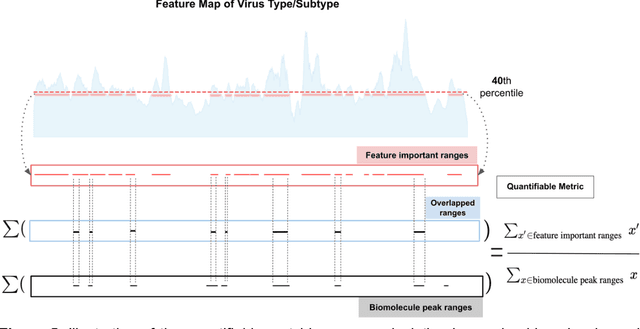
Abstract:Rapid identification of newly emerging or circulating viruses is an important first step toward managing the public health response to potential outbreaks. A portable virus capture device coupled with label-free Raman Spectroscopy holds the promise of fast detection by rapidly obtaining the Raman signature of a virus followed by a machine learning approach applied to recognize the virus based on its Raman spectrum, which is used as a fingerprint. We present such a machine learning approach for analyzing Raman spectra of human and avian viruses. A Convolutional Neural Network (CNN) classifier specifically designed for spectral data achieves very high accuracy for a variety of virus type or subtype identification tasks. In particular, it achieves 99% accuracy for classifying influenza virus type A vs. type B, 96% accuracy for classifying four subtypes of influenza A, 95% accuracy for differentiating enveloped and non-enveloped viruses, and 99% accuracy for differentiating avian coronavirus (infectious bronchitis virus, IBV) from other avian viruses. Furthermore, interpretation of neural net responses in the trained CNN model using a full-gradient algorithm highlights Raman spectral ranges that are most important to virus identification. By correlating ML-selected salient Raman ranges with the signature ranges of known biomolecules and chemical functional groups (for example, amide, amino acid, carboxylic acid), we verify that our ML model effectively recognizes the Raman signatures of proteins, lipids and other vital functional groups present in different viruses and uses a weighted combination of these signatures to identify viruses.
* 23 pages, 8 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge