R. Joe Stanley
Increasing Melanoma Diagnostic Confidence: Forcing the Convolutional Network to Learn from the Lesion
May 16, 2023Abstract:Deep learning implemented with convolutional network architectures can exceed specialists' diagnostic accuracy. However, whole-image deep learning trained on a given dataset may not generalize to other datasets. The problem arises because extra-lesional features - ruler marks, ink marks, and other melanoma correlates - may serve as information leaks. These extra-lesional features, discoverable by heat maps, degrade melanoma diagnostic performance and cause techniques learned on one data set to fail to generalize. We propose a novel technique to improve melanoma recognition by an EfficientNet model. The model trains the network to detect the lesion and learn features from the detected lesion. A generalizable elliptical segmentation model for lesions was developed, with an ellipse enclosing a lesion and the ellipse enclosed by an extended rectangle (bounding box). The minimal bounding box was extended by 20% to allow some background around the lesion. The publicly available International Skin Imaging Collaboration (ISIC) 2020 skin lesion image dataset was used to evaluate the effectiveness of the proposed method. Our test results show that the proposed method improved diagnostic accuracy by increasing the mean area under receiver operating characteristic curve (mean AUC) score from 0.9 to 0.922. Additionally, correctly diagnosed scores are also improved, providing better separation of scores, thereby increasing melanoma diagnostic confidence. The proposed lesion-focused convolutional technique warrants further study.
Feature based Sequential Classifier with Attention Mechanism
Jul 22, 2020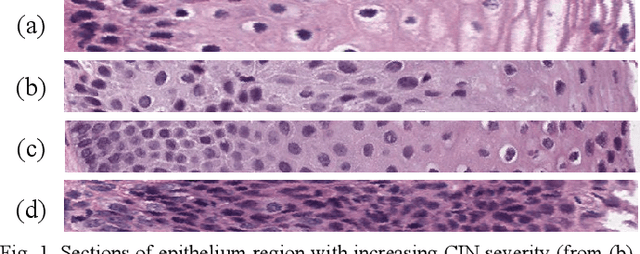
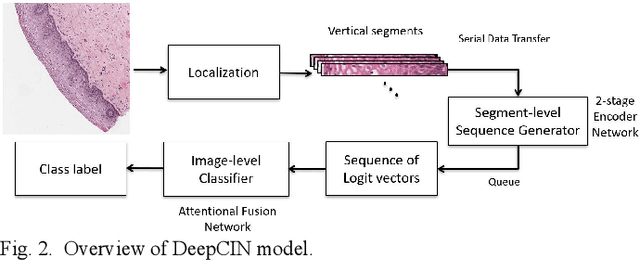
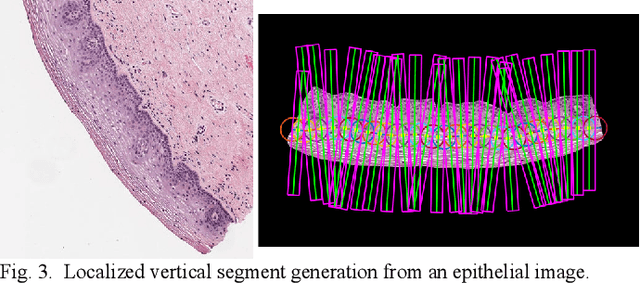
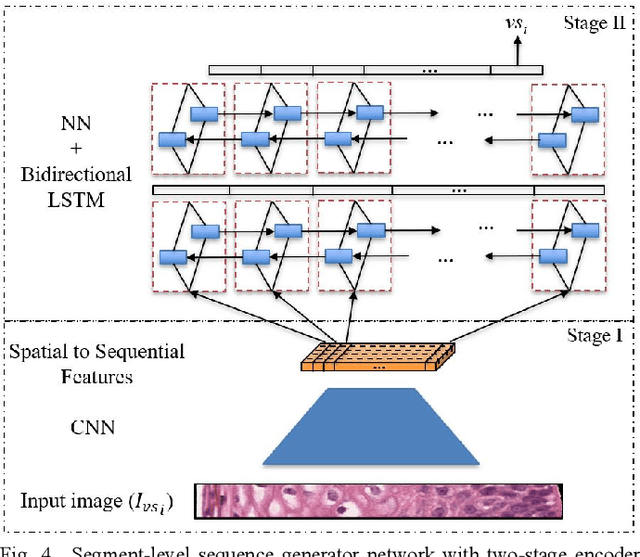
Abstract:Cervical cancer is one of the deadliest cancers affecting women globally. Cervical intraepithelial neoplasia (CIN) assessment using histopathological examination of cervical biopsy slides is subject to interobserver variability. Automated processing of digitized histopathology slides has the potential for more accurate classification for CIN grades from normal to increasing grades of pre-malignancy: CIN1, CIN2 and CIN3. Cervix disease is generally understood to progress from the bottom (basement membrane) to the top of the epithelium. To model this relationship of disease severity to spatial distribution of abnormalities, we propose a network pipeline, DeepCIN, to analyze high-resolution epithelium images (manually extracted from whole-slide images) hierarchically by focusing on localized vertical regions and fusing this local information for determining Normal/CIN classification. The pipeline contains two classifier networks: 1) a cross-sectional, vertical segment-level sequence generator (two-stage encoder model) is trained using weak supervision to generate feature sequences from the vertical segments to preserve the bottom-to-top feature relationships in the epithelium image data; 2) an attention-based fusion network image-level classifier predicting the final CIN grade by merging vertical segment sequences. The model produces the CIN classification results and also determines the vertical segment contributions to CIN grade prediction. Experiments show that DeepCIN achieves pathologist-level CIN classification accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge