Sivaramakrishnan Rajaraman
Uncovering the effects of model initialization on deep model generalization: A study with adult and pediatric Chest X-ray images
Sep 20, 2023Abstract:Model initialization techniques are vital for improving the performance and reliability of deep learning models in medical computer vision applications. While much literature exists on non-medical images, the impacts on medical images, particularly chest X-rays (CXRs) are less understood. Addressing this gap, our study explores three deep model initialization techniques: Cold-start, Warm-start, and Shrink and Perturb start, focusing on adult and pediatric populations. We specifically focus on scenarios with periodically arriving data for training, thereby embracing the real-world scenarios of ongoing data influx and the need for model updates. We evaluate these models for generalizability against external adult and pediatric CXR datasets. We also propose novel ensemble methods: F-score-weighted Sequential Least-Squares Quadratic Programming (F-SLSQP) and Attention-Guided Ensembles with Learnable Fuzzy Softmax to aggregate weight parameters from multiple models to capitalize on their collective knowledge and complementary representations. We perform statistical significance tests with 95% confidence intervals and p-values to analyze model performance. Our evaluations indicate models initialized with ImageNet-pre-trained weights demonstrate superior generalizability over randomly initialized counterparts, contradicting some findings for non-medical images. Notably, ImageNet-pretrained models exhibit consistent performance during internal and external testing across different training scenarios. Weight-level ensembles of these models show significantly higher recall (p<0.05) during testing compared to individual models. Thus, our study accentuates the benefits of ImageNet-pretrained weight initialization, especially when used with weight-level ensembles, for creating robust and generalizable deep learning solutions.
Semantically Redundant Training Data Removal and Deep Model Classification Performance: A Study with Chest X-rays
Sep 18, 2023Abstract:Deep learning (DL) has demonstrated its innate capacity to independently learn hierarchical features from complex and multi-dimensional data. A common understanding is that its performance scales up with the amount of training data. Another data attribute is the inherent variety. It follows, therefore, that semantic redundancy, which is the presence of similar or repetitive information, would tend to lower performance and limit generalizability to unseen data. In medical imaging data, semantic redundancy can occur due to the presence of multiple images that have highly similar presentations for the disease of interest. Further, the common use of augmentation methods to generate variety in DL training may be limiting performance when applied to semantically redundant data. We propose an entropy-based sample scoring approach to identify and remove semantically redundant training data. We demonstrate using the publicly available NIH chest X-ray dataset that the model trained on the resulting informative subset of training data significantly outperforms the model trained on the full training set, during both internal (recall: 0.7164 vs 0.6597, p<0.05) and external testing (recall: 0.3185 vs 0.2589, p<0.05). Our findings emphasize the importance of information-oriented training sample selection as opposed to the conventional practice of using all available training data.
Does image resolution impact chest X-ray based fine-grained Tuberculosis-consistent lesion segmentation?
Jan 10, 2023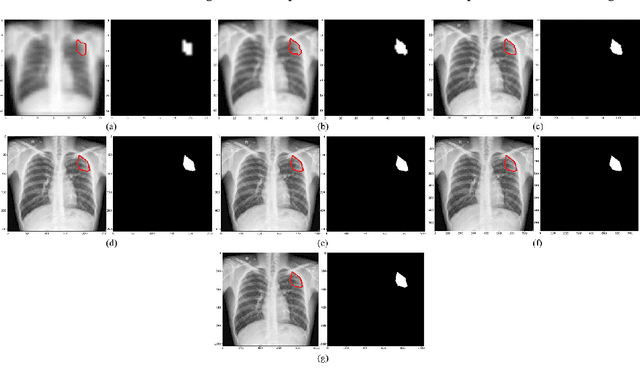
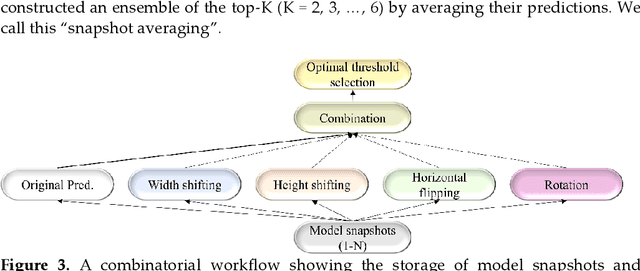
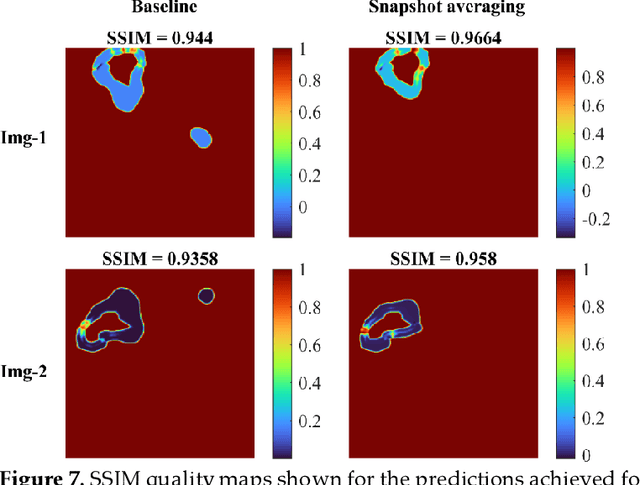
Abstract:Deep learning (DL) models are becoming state-of-the-art in segmenting anatomical and disease regions of interest (ROIs) in medical images, particularly chest X-rays (CXRs). However, these models are reportedly trained on reduced image resolutions citing reasons for the lack of computational resources. Literature is sparse considering identifying the optimal image resolution to train these models for the task under study, particularly considering segmentation of Tuberculosis (TB)-consistent lesions in CXRs. In this study, we used the (i) Shenzhen TB CXR dataset, investigated performance gains achieved through training an Inception-V3-based UNet model using various image/mask resolutions with/without lung ROI cropping and aspect ratio adjustments, and (ii) identified the optimal image resolution through extensive empirical evaluations to improve TB-consistent lesion segmentation performance. We proposed a combinatorial approach consisting of storing model snapshots, optimizing test-time augmentation (TTA) methods, and selecting the optimal segmentation threshold to further improve performance at the optimal resolution. We emphasize that (i) higher image resolutions are not always necessary and (ii) identifying the optimal image resolution is indispensable to achieve superior performance for the task under study.
Generalizability of Deep Adult Lung Segmentation Models to the Pediatric Population: A Retrospective Study
Nov 04, 2022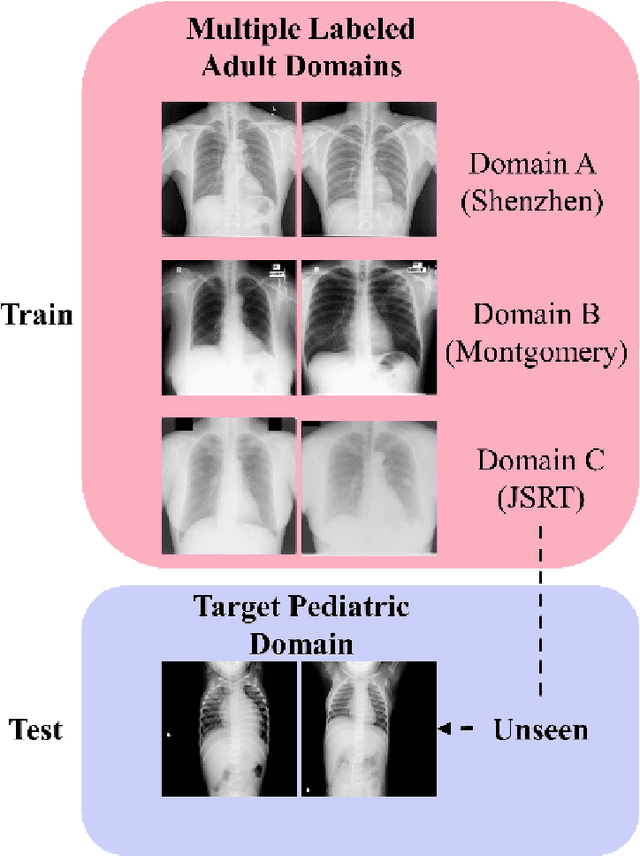

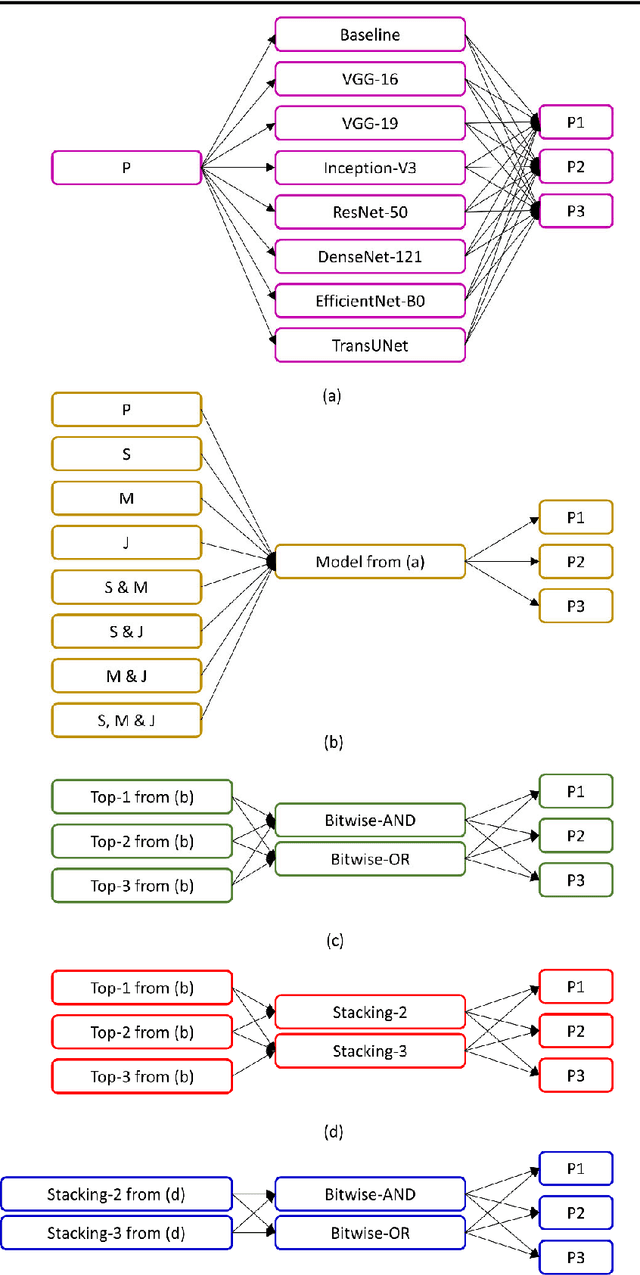

Abstract:Lung segmentation in chest X-rays (CXRs) is an important prerequisite for improving the specificity of diagnoses of cardiopulmonary diseases in a clinical decision support system. Current deep learning (DL) models for lung segmentation are trained and evaluated on CXR datasets in which the radiographic projections are captured predominantly from the adult population. However, the shape of the lungs is reported to be significantly different for pediatrics across the developmental stages from infancy to adulthood. This might result in age-related data domain shifts that would adversely impact lung segmentation performance when the models trained on the adult population are deployed for pediatric lung segmentation. In this work, our goal is to analyze the generalizability of deep adult lung segmentation models to the pediatric population and improve performance through a systematic combinatorial approach consisting of CXR modality-specific weight initializations, stacked generalization, and an ensemble of the stacked generalization models. Novel evaluation metrics consisting of Mean Lung Contour Distance and Average Hash Score are proposed in addition to the Multi-scale Structural Similarity Index Measure, Intersection of Union, and Dice metrics to evaluate segmentation performance. We observed a significant improvement (p < 0.05) in cross-domain generalization through our combinatorial approach. This study could serve as a paradigm to analyze the cross-domain generalizability of deep segmentation models for other medical imaging modalities and applications.
Deep ensemble learning for segmenting tuberculosis-consistent manifestations in chest radiographs
Jun 13, 2022Abstract:Automated segmentation of tuberculosis (TB)-consistent lesions in chest X-rays (CXRs) using deep learning (DL) methods can help reduce radiologist effort, supplement clinical decision-making, and potentially result in improved patient treatment. The majority of works in the literature discuss training automatic segmentation models using coarse bounding box annotations. However, the granularity of the bounding box annotation could result in the inclusion of a considerable fraction of false positives and negatives at the pixel level that may adversely impact overall semantic segmentation performance. This study (i) evaluates the benefits of using fine-grained annotations of TB-consistent lesions and (ii) trains and constructs ensembles of the variants of U-Net models for semantically segmenting TB-consistent lesions in both original and bone-suppressed frontal CXRs. We evaluated segmentation performance using several ensemble methods such as bitwise AND, bitwise-OR, bitwise-MAX, and stacking. We observed that the stacking ensemble demonstrated superior segmentation performance (Dice score: 0.5743, 95% confidence interval: (0.4055,0.7431)) compared to the individual constituent models and other ensemble methods. To the best of our knowledge, this is the first study to apply ensemble learning to improve fine-grained TB-consistent lesion segmentation performance.
A bone suppression model ensemble to improve COVID-19 detection in chest X-rays
Nov 05, 2021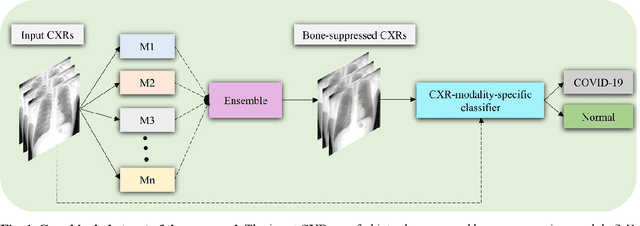
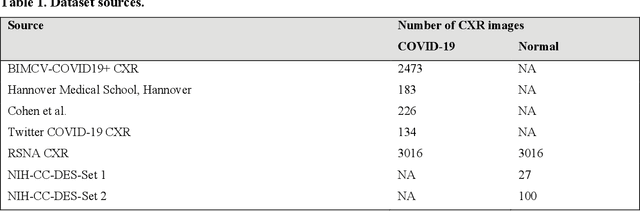
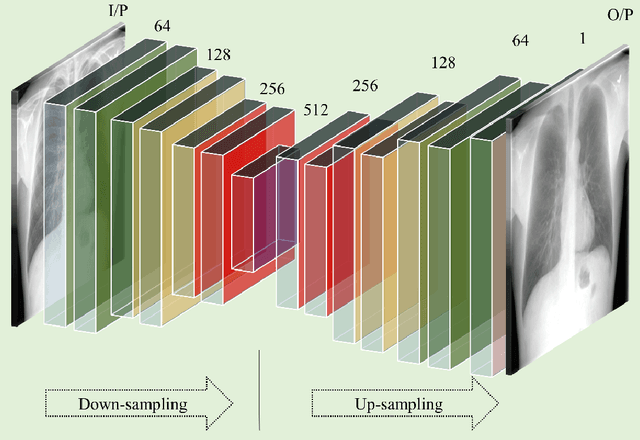
Abstract:Chest X-ray (CXR) is a widely performed radiology examination that helps to detect abnormalities in the tissues and organs in the thoracic cavity. Detecting pulmonary abnormalities like COVID-19 may become difficult due to that they are obscured by the presence of bony structures like the ribs and the clavicles, thereby resulting in screening/diagnostic misinterpretations. Automated bone suppression methods would help suppress these bony structures and increase soft tissue visibility. In this study, we propose to build an ensemble of convolutional neural network models to suppress bones in frontal CXRs, improve classification performance, and reduce interpretation errors related to COVID-19 detection. The ensemble is constructed by (i) measuring the multi-scale structural similarity index (MS-SSIM) score between the sub-blocks of the bone-suppressed image predicted by each of the top-3 performing bone-suppression models and the corresponding sub-blocks of its respective ground truth soft-tissue image, and (ii) performing a majority voting of the MS-SSIM score computed in each sub-block to identify the sub-block with the maximum MS-SSIM score and use it in constructing the final bone-suppressed image. We empirically determine the sub-block size that delivers superior bone suppression performance. It is observed that the bone suppression model ensemble outperformed the individual models in terms of MS-SSIM and other metrics. A CXR modality-specific classification model is retrained and evaluated on the non-bone-suppressed and bone-suppressed images to classify them as showing normal lungs or other COVID-19-like manifestations. We observed that the bone-suppressed model training significantly outperformed the model trained on non-bone-suppressed images toward detecting COVID-19 manifestations.
Does deep learning model calibration improve performance in class-imbalanced medical image classification?
Oct 11, 2021
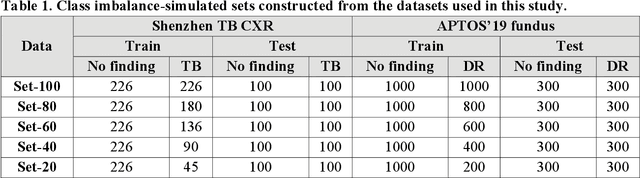
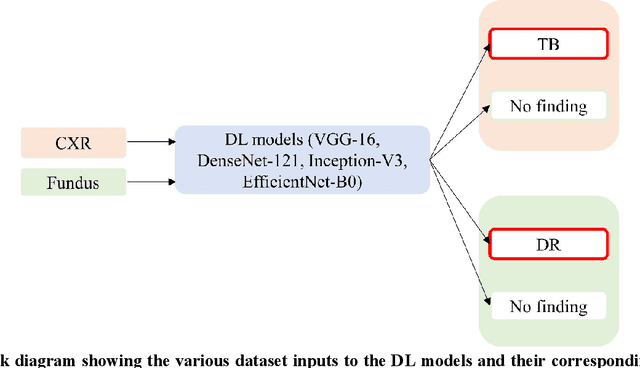
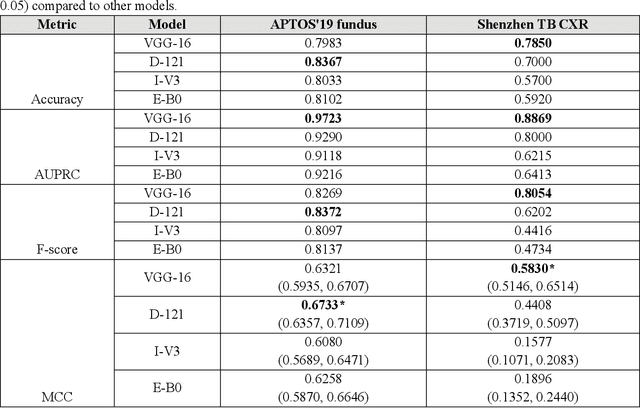
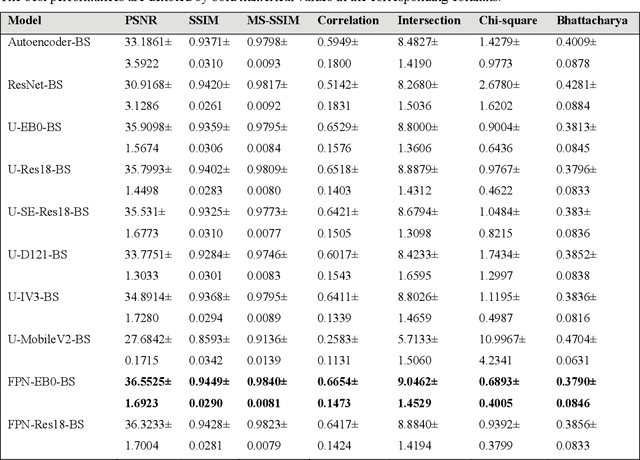
Abstract:In medical image classification tasks, it is common to find that the number of normal samples far exceeds the number of abnormal samples. In such class-imbalanced situations, reliable training of deep neural networks continues to be a major challenge. Under these circumstances, the predicted class probabilities may be biased toward the majority class. Calibration has been suggested to alleviate some of these effects. However, there is insufficient analysis explaining when and whether calibrating a model would be beneficial in improving performance. In this study, we perform a systematic analysis of the effect of model calibration on its performance on two medical image modalities, namely, chest X-rays and fundus images, using various deep learning classifier backbones. For this, we study the following variations: (i) the degree of imbalances in the dataset used for training; (ii) calibration methods; and (iii) two classification thresholds, namely, default decision threshold of 0.5, and optimal threshold from precision-recall curves. Our results indicate that at the default operating threshold of 0.5, the performance achieved through calibration is significantly superior (p < 0.05) to using uncalibrated probabilities. However, at the PR-guided threshold, these gains are not significantly different (p > 0.05). This finding holds for both image modalities and at varying degrees of imbalance.
Multi-loss ensemble deep learning for chest X-ray classification
Sep 29, 2021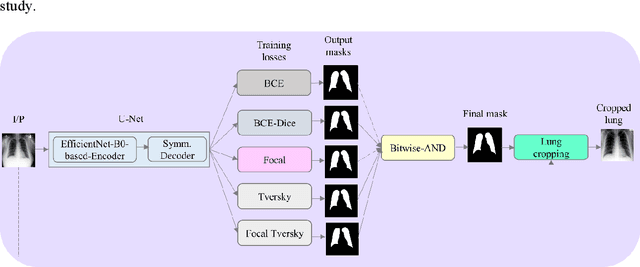
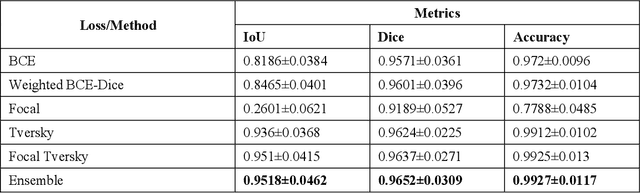
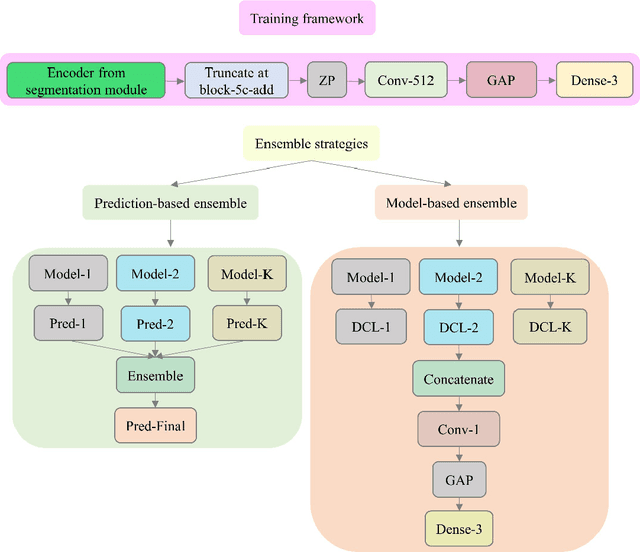
Abstract:Class imbalance is common in medical image classification tasks, where the number of abnormal samples is fewer than the number of normal samples. The difficulty of imbalanced classification is compounded by other issues such as the size and distribution of the dataset. Reliable training of deep neural networks continues to be a major challenge in such class-imbalanced conditions. The loss function used to train the deep neural networks highly impact the performance of both balanced and imbalanced tasks. Currently, the cross-entropy loss remains the de-facto loss function for balanced and imbalanced classification tasks. This loss, however, asserts equal learning to all classes, leading to the classification of most samples as the majority normal class. To provide a critical analysis of different loss functions and identify those suitable for class-imbalanced classification, we benchmark various state-of-the-art loss functions and propose novel loss functions to train a DL model and analyze its performance in a multiclass classification setting that classifies pediatric chest X-rays as showing normal lungs, bacterial pneumonia, or viral pneumonia manifestations. We also construct prediction-level and model-level ensembles of the models that are trained with various loss functions to improve classification performance. We performed localization studies to interpret model behavior to ensure that the individual models and their ensembles precisely learned the regions of interest showing disease manifestations to classify the chest X-rays to their respective categories.
Chest X-Ray Bone Suppression for Improving Classification of Tuberculosis-Consistent Findings
May 07, 2021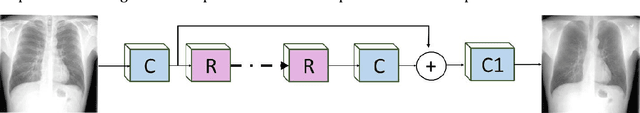
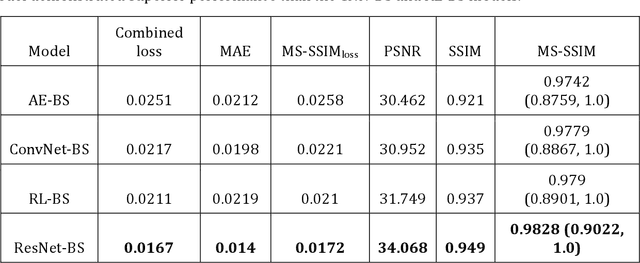
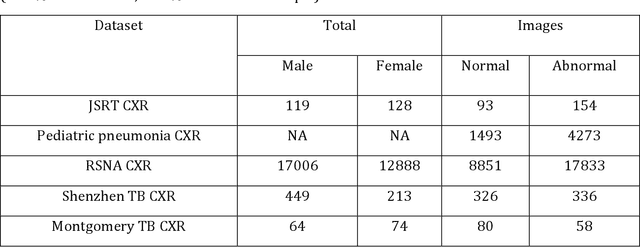
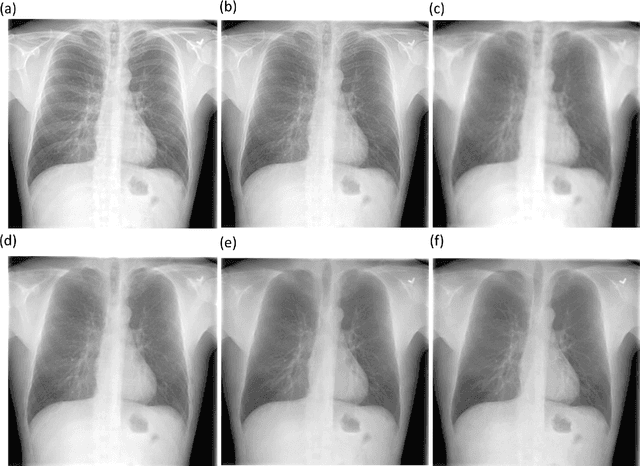
Abstract:Chest X-rays are the most commonly performed diagnostic examination to detect cardiopulmonary abnormalities. However, the presence of bony structures such as ribs and clavicles can obscure subtle abnormalities, resulting in diagnostic errors. This study aims to build a deep learning-based bone suppression model that identifies and removes these occluding bony structures in frontal CXRs to assist in reducing errors in radiological interpretation, including DL workflows, related to detecting manifestations consistent with tuberculosis (TB). Several bone suppression models with various deep architectures are trained and optimized using the proposed combined loss function and their performances are evaluated in a cross-institutional test setting. The best-performing model is used to suppress bones in the publicly available Shenzhen and Montgomery TB CXR collections. A VGG-16 model is pretrained on a large collection of publicly available CXRs. The CXR-pretrained model is then fine-tuned individually on the non-bone-suppressed and bone-suppressed CXRs of Shenzhen and Montgomery TB CXR collections to classify them as showing normal lungs or TB manifestations. The performances of these models are compared using several performance metrics, analyzed for statistical significance, and their predictions are qualitatively interpreted through class-selective relevance maps. It is observed that the models trained on bone-suppressed CXRs significantly outperformed (p<0.05) the models trained on the non-bone-suppressed CXRs. Models trained on bone-suppressed CXRs improved detection of TB-consistent findings and resulted in compact clustering of the data points in the feature space signifying that bone suppression improved the model sensitivity toward TB classification.
Training custom modality-specific U-Net models with weak localizations for improved Tuberculosis segmentation and localization
Mar 01, 2021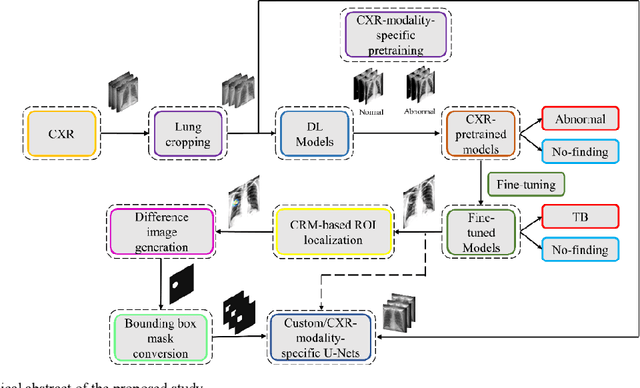
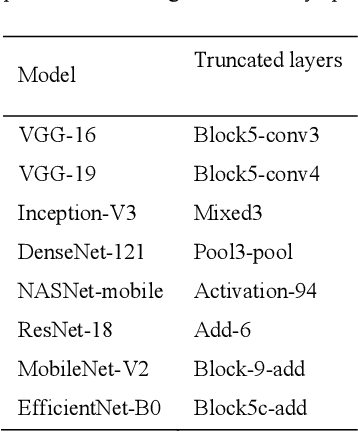
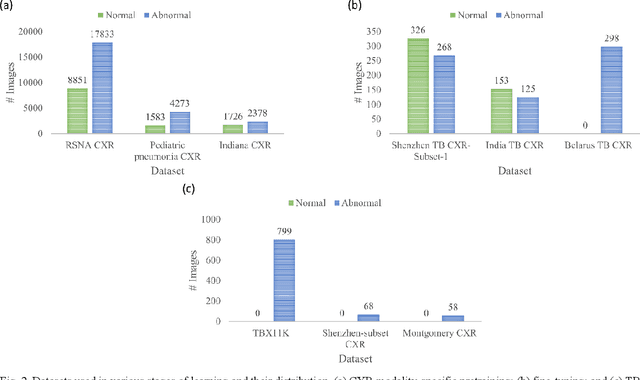
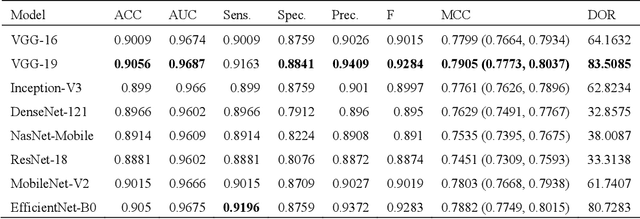
Abstract:Deep learning (DL) has drawn tremendous attention in object localization and recognition for both natural and medical images. U-Net segmentation models have demonstrated superior performance compared to conventional hand-crafted feature-based methods. Medical image modality-specific DL models are better at transferring domain knowledge to a relevant target task than those that are pretrained on stock photography images. This helps improve model adaptation, generalization, and class-specific region of interest (ROI) localization. In this study, we train chest X-ray (CXR) modality-specific U-Nets and other state-of-the-art U-Net models for semantic segmentation of tuberculosis (TB)-consistent findings. Automated segmentation of such manifestations could help radiologists reduce errors and supplement decision-making while improving patient care and productivity. Our approach uses the publicly available TBX11K CXR dataset with weak TB annotations, typically provided as bounding boxes, to train a set of U-Net models. Next, we improve the results by augmenting the training data with weak localizations, post-processed into an ROI mask, from a DL classifier that is trained to classify CXRs as showing normal lungs or suspected TB manifestations. Test data are individually derived from the TBX11K CXR training distribution and other cross-institutional collections including the Shenzhen TB and Montgomery TB CXR datasets. We observe that our augmented training strategy helped the CXR modality-specific U-Net models achieve superior performance with test data derived from the TBX11K CXR training distribution as well as from cross-institutional collections (p < 0.05).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge