Les Folio
Improving Radiology Report Conciseness and Structure via Local Large Language Models
Nov 06, 2024Abstract:In this study, we aim to enhance radiology reporting by improving both the conciseness and structured organization of findings (also referred to as templating), specifically by organizing information according to anatomical regions. This structured approach allows physicians to locate relevant information quickly, increasing the report's utility. We utilize Large Language Models (LLMs) such as Mixtral, Mistral, and Llama to generate concise, well-structured reports. Among these, we primarily focus on the Mixtral model due to its superior adherence to specific formatting requirements compared to other models. To maintain data security and privacy, we run these LLMs locally behind our institution's firewall. We leverage the LangChain framework and apply five distinct prompting strategies to enforce a consistent structure in radiology reports, aiming to eliminate extraneous language and achieve a high level of conciseness. We also introduce a novel metric, the Conciseness Percentage (CP) score, to evaluate report brevity. Our dataset comprises 814 radiology reports authored by seven board-certified body radiologists at our cancer center. In evaluating the different prompting methods, we discovered that the most effective approach for generating concise, well-structured reports involves first instructing the LLM to condense the report, followed by a prompt to structure the content according to specific guidelines. We assessed all prompting strategies based on their ability to handle formatting issues, reduce report length, and adhere to formatting instructions. Our findings demonstrate that open-source, locally deployed LLMs can significantly improve radiology report conciseness and structure while conforming to specified formatting standards.
Chest X-Ray Bone Suppression for Improving Classification of Tuberculosis-Consistent Findings
May 07, 2021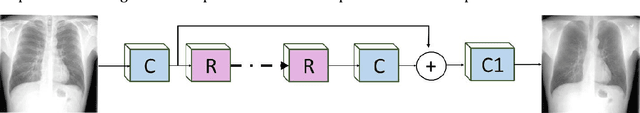
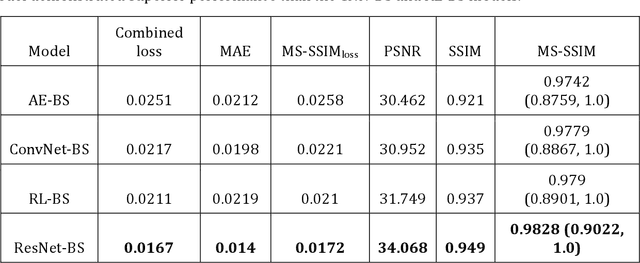
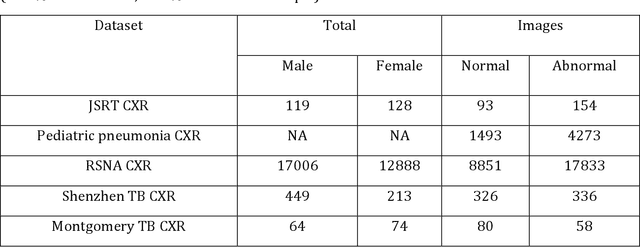
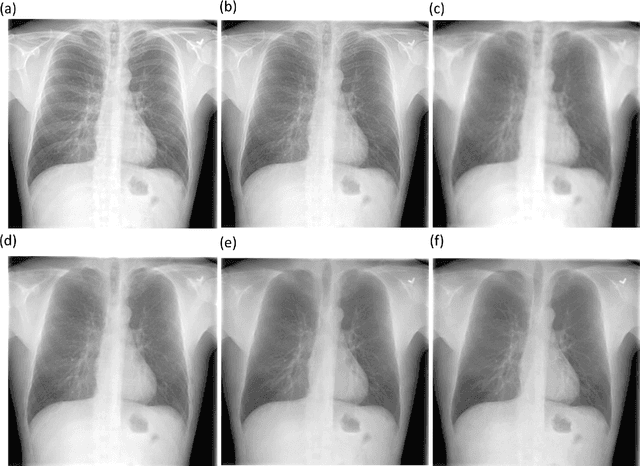
Abstract:Chest X-rays are the most commonly performed diagnostic examination to detect cardiopulmonary abnormalities. However, the presence of bony structures such as ribs and clavicles can obscure subtle abnormalities, resulting in diagnostic errors. This study aims to build a deep learning-based bone suppression model that identifies and removes these occluding bony structures in frontal CXRs to assist in reducing errors in radiological interpretation, including DL workflows, related to detecting manifestations consistent with tuberculosis (TB). Several bone suppression models with various deep architectures are trained and optimized using the proposed combined loss function and their performances are evaluated in a cross-institutional test setting. The best-performing model is used to suppress bones in the publicly available Shenzhen and Montgomery TB CXR collections. A VGG-16 model is pretrained on a large collection of publicly available CXRs. The CXR-pretrained model is then fine-tuned individually on the non-bone-suppressed and bone-suppressed CXRs of Shenzhen and Montgomery TB CXR collections to classify them as showing normal lungs or TB manifestations. The performances of these models are compared using several performance metrics, analyzed for statistical significance, and their predictions are qualitatively interpreted through class-selective relevance maps. It is observed that the models trained on bone-suppressed CXRs significantly outperformed (p<0.05) the models trained on the non-bone-suppressed CXRs. Models trained on bone-suppressed CXRs improved detection of TB-consistent findings and resulted in compact clustering of the data points in the feature space signifying that bone suppression improved the model sensitivity toward TB classification.
Training custom modality-specific U-Net models with weak localizations for improved Tuberculosis segmentation and localization
Mar 01, 2021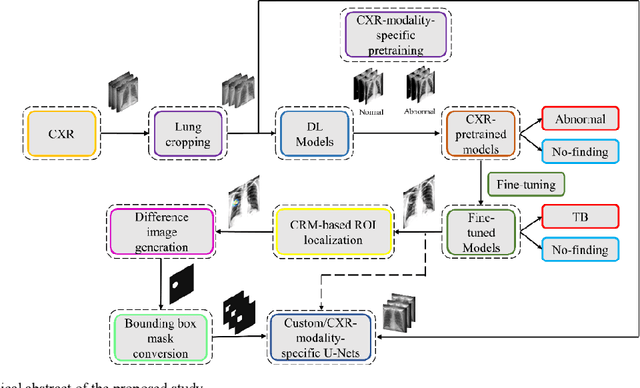
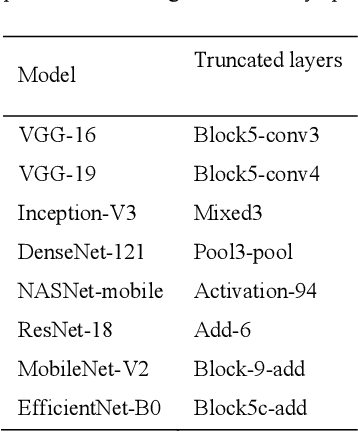
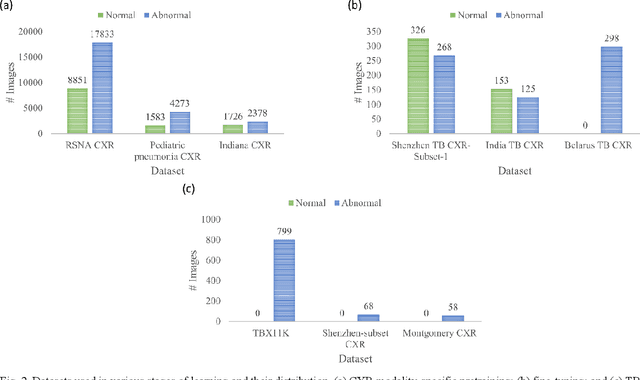
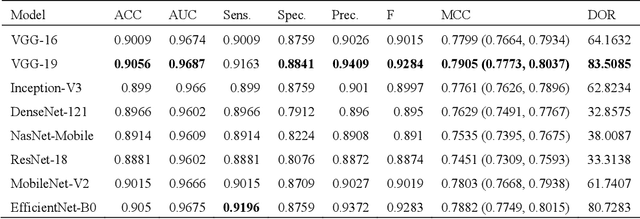
Abstract:Deep learning (DL) has drawn tremendous attention in object localization and recognition for both natural and medical images. U-Net segmentation models have demonstrated superior performance compared to conventional hand-crafted feature-based methods. Medical image modality-specific DL models are better at transferring domain knowledge to a relevant target task than those that are pretrained on stock photography images. This helps improve model adaptation, generalization, and class-specific region of interest (ROI) localization. In this study, we train chest X-ray (CXR) modality-specific U-Nets and other state-of-the-art U-Net models for semantic segmentation of tuberculosis (TB)-consistent findings. Automated segmentation of such manifestations could help radiologists reduce errors and supplement decision-making while improving patient care and productivity. Our approach uses the publicly available TBX11K CXR dataset with weak TB annotations, typically provided as bounding boxes, to train a set of U-Net models. Next, we improve the results by augmenting the training data with weak localizations, post-processed into an ROI mask, from a DL classifier that is trained to classify CXRs as showing normal lungs or suspected TB manifestations. Test data are individually derived from the TBX11K CXR training distribution and other cross-institutional collections including the Shenzhen TB and Montgomery TB CXR datasets. We observe that our augmented training strategy helped the CXR modality-specific U-Net models achieve superior performance with test data derived from the TBX11K CXR training distribution as well as from cross-institutional collections (p < 0.05).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge