Zhenyu Bi
DEBATE, TRAIN, EVOLVE: Self Evolution of Language Model Reasoning
May 21, 2025Abstract:Large language models (LLMs) have improved significantly in their reasoning through extensive training on massive datasets. However, relying solely on additional data for improvement is becoming increasingly impractical, highlighting the need for models to autonomously enhance their reasoning without external supervision. In this paper, we propose Debate, Train, Evolve (DTE), a novel ground truth-free training framework that uses multi-agent debate traces to evolve a single language model. We also introduce a new prompting strategy Reflect-Critique-Refine, to improve debate quality by explicitly instructing agents to critique and refine their reasoning. Extensive evaluations on five reasoning benchmarks with six open-weight models show that our DTE framework achieve substantial improvements, with an average accuracy gain of 8.92% on the challenging GSM-PLUS dataset. Furthermore, we observe strong cross-domain generalization, with an average accuracy gain of 5.8% on all other benchmarks, suggesting that our method captures general reasoning capabilities.
Population Aware Diffusion for Time Series Generation
Jan 01, 2025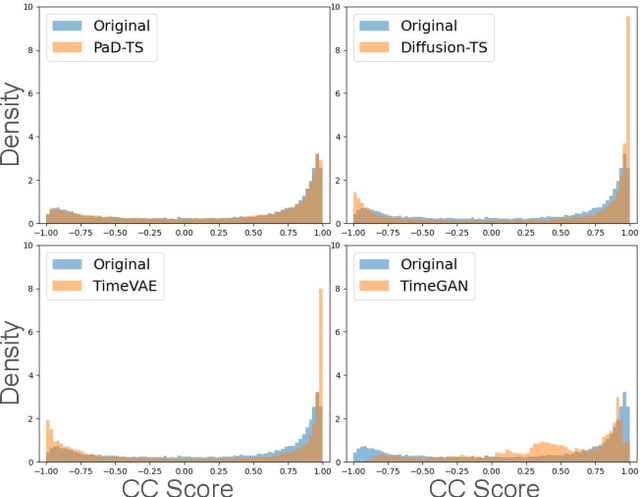



Abstract:Diffusion models have shown promising ability in generating high-quality time series (TS) data. Despite the initial success, existing works mostly focus on the authenticity of data at the individual level, but pay less attention to preserving the population-level properties on the entire dataset. Such population-level properties include value distributions for each dimension and distributions of certain functional dependencies (e.g., cross-correlation, CC) between different dimensions. For instance, when generating house energy consumption TS data, the value distributions of the outside temperature and the kitchen temperature should be preserved, as well as the distribution of CC between them. Preserving such TS population-level properties is critical in maintaining the statistical insights of the datasets, mitigating model bias, and augmenting downstream tasks like TS prediction. Yet, it is often overlooked by existing models. Hence, data generated by existing models often bear distribution shifts from the original data. We propose Population-aware Diffusion for Time Series (PaD-TS), a new TS generation model that better preserves the population-level properties. The key novelties of PaD-TS include 1) a new training method explicitly incorporating TS population-level property preservation, and 2) a new dual-channel encoder model architecture that better captures the TS data structure. Empirical results in major benchmark datasets show that PaD-TS can improve the average CC distribution shift score between real and synthetic data by 5.9x while maintaining a performance comparable to state-of-the-art models on individual-level authenticity.
STOC-TOT: Stochastic Tree-of-Thought with Constrained Decoding for Complex Reasoning in Multi-Hop Question Answering
Jul 04, 2024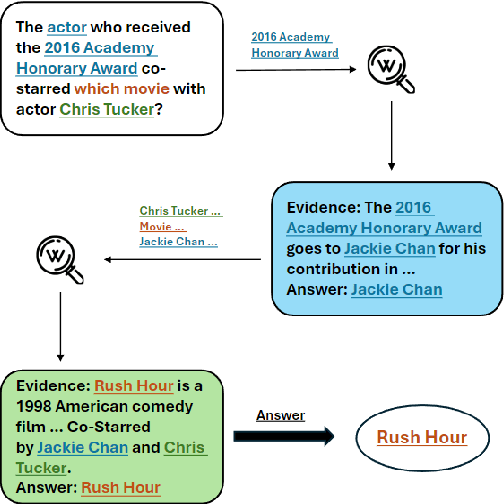
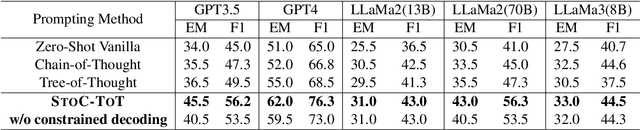
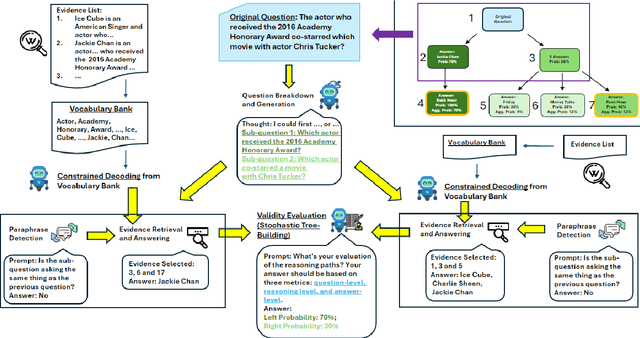
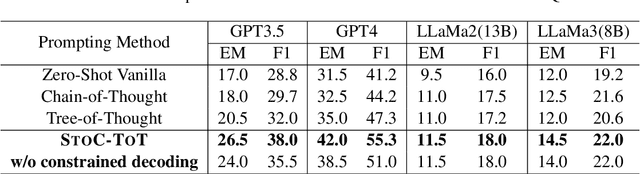
Abstract:Multi-hop question answering (MHQA) requires a model to retrieve and integrate information from multiple passages to answer a complex question. Recent systems leverage the power of large language models and integrate evidence retrieval with reasoning prompts (e.g., chain-of-thought reasoning) for the MHQA task. However, the complexities in the question types (bridge v.s. comparison questions) and the reasoning types (sequential v.s. parallel reasonings) require more novel and fine-grained prompting methods to enhance the performance of MHQA under the zero-shot setting. In this paper, we propose STOC-TOT, a stochastic tree-of-thought reasoning prompting method with constrained decoding for MHQA and conduct a detailed comparison with other reasoning prompts on different question types and reasoning types. Specifically, we construct a tree-like reasoning structure by prompting the model to break down the original question into smaller sub-questions to form different reasoning paths. In addition, we prompt the model to provide a probability estimation for each reasoning path at each reasoning step. At answer time, we conduct constrained decoding on the model to generate more grounded answers and reduce hallucination. Experiments comparing STOC-TOT with two MHQA datasets and five large language models showed that our framework outperforms other reasoning prompts by a significant margin.
AI for Biomedicine in the Era of Large Language Models
Mar 23, 2024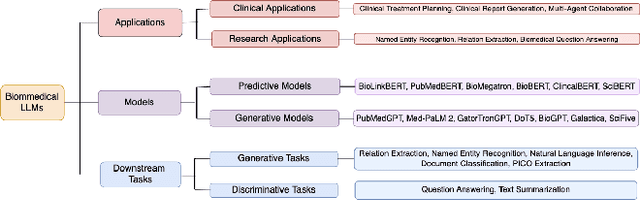
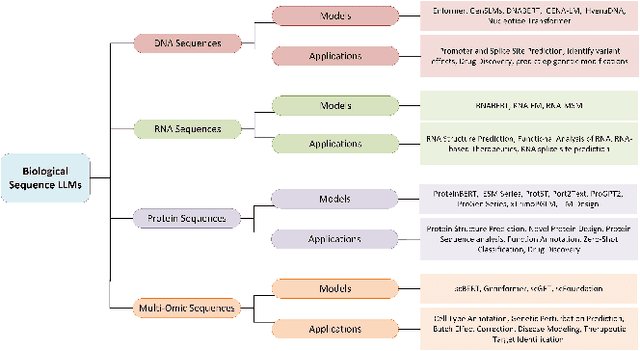
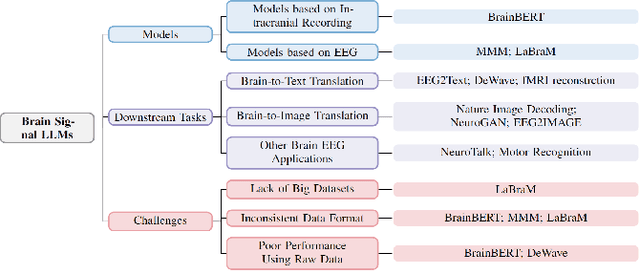
Abstract:The capabilities of AI for biomedicine span a wide spectrum, from the atomic level, where it solves partial differential equations for quantum systems, to the molecular level, predicting chemical or protein structures, and further extending to societal predictions like infectious disease outbreaks. Recent advancements in large language models, exemplified by models like ChatGPT, have showcased significant prowess in natural language tasks, such as translating languages, constructing chatbots, and answering questions. When we consider biomedical data, we observe a resemblance to natural language in terms of sequences: biomedical literature and health records presented as text, biological sequences or sequencing data arranged in sequences, or sensor data like brain signals as time series. The question arises: Can we harness the potential of recent large language models to drive biomedical knowledge discoveries? In this survey, we will explore the application of large language models to three crucial categories of biomedical data: 1) textual data, 2) biological sequences, and 3) brain signals. Furthermore, we will delve into large language model challenges in biomedical research, including ensuring trustworthiness, achieving personalization, and adapting to multi-modal data representation
UCPhrase: Unsupervised Context-aware Quality Phrase Tagging
May 28, 2021
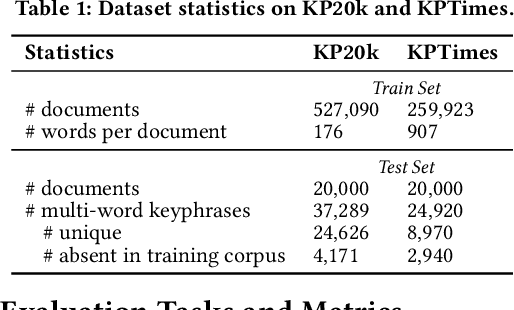
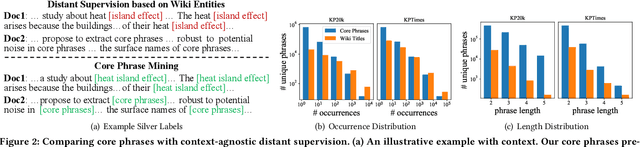
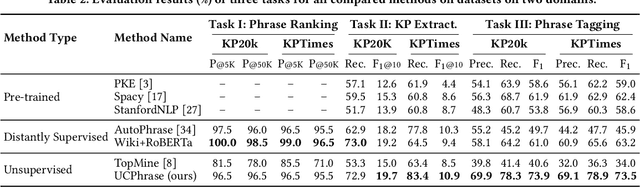
Abstract:Identifying and understanding quality phrases from context is a fundamental task in text mining. The most challenging part of this task arguably lies in uncommon, emerging, and domain-specific phrases. The infrequent nature of these phrases significantly hurts the performance of phrase mining methods that rely on sufficient phrase occurrences in the input corpus. Context-aware tagging models, though not restricted by frequency, heavily rely on domain experts for either massive sentence-level gold labels or handcrafted gazetteers. In this work, we propose UCPhrase, a novel unsupervised context-aware quality phrase tagger. Specifically, we induce high-quality phrase spans as silver labels from consistently co-occurring word sequences within each document. Compared with typical context-agnostic distant supervision based on existing knowledge bases (KBs), our silver labels root deeply in the input domain and context, thus having unique advantages in preserving contextual completeness and capturing emerging, out-of-KB phrases. Training a conventional neural tagger based on silver labels usually faces the risk of overfitting phrase surface names. Alternatively, we observe that the contextualized attention maps generated from a transformer-based neural language model effectively reveal the connections between words in a surface-agnostic way. Therefore, we pair such attention maps with the silver labels to train a lightweight span prediction model, which can be applied to new input to recognize (unseen) quality phrases regardless of their surface names or frequency. Thorough experiments on various tasks and datasets, including corpus-level phrase ranking, document-level keyphrase extraction, and sentence-level phrase tagging, demonstrate the superiority of our design over state-of-the-art pre-trained, unsupervised, and distantly supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge