Zhenghua Xu
Language Drift in Multilingual Retrieval-Augmented Generation: Characterization and Decoding-Time Mitigation
Nov 13, 2025



Abstract:Multilingual Retrieval-Augmented Generation (RAG) enables large language models (LLMs) to perform knowledge-intensive tasks in multilingual settings by leveraging retrieved documents as external evidence. However, when the retrieved evidence differs in language from the user query and in-context exemplars, the model often exhibits language drift by generating responses in an unintended language. This phenomenon is especially pronounced during reasoning-intensive decoding, such as Chain-of-Thought (CoT) generation, where intermediate steps introduce further language instability. In this paper, we systematically study output language drift in multilingual RAG across multiple datasets, languages, and LLM backbones. Our controlled experiments reveal that the drift results not from comprehension failure but from decoder-level collapse, where dominant token distributions and high-frequency English patterns dominate the intended generation language. We further observe that English serves as a semantic attractor under cross-lingual conditions, emerging as both the strongest interference source and the most frequent fallback language. To mitigate this, we propose Soft Constrained Decoding (SCD), a lightweight, training-free decoding strategy that gently steers generation toward the target language by penalizing non-target-language tokens. SCD is model-agnostic and can be applied to any generation algorithm without modifying the architecture or requiring additional data. Experiments across three multilingual datasets and multiple typologically diverse languages show that SCD consistently improves language alignment and task performance, providing an effective and generalizable solution in multilingual RAG.
Modeling Uncertainty Trends for Timely Retrieval in Dynamic RAG
Nov 13, 2025Abstract:Dynamic retrieval-augmented generation (RAG) allows large language models (LLMs) to fetch external knowledge on demand, offering greater adaptability than static RAG. A central challenge in this setting lies in determining the optimal timing for retrieval. Existing methods often trigger retrieval based on low token-level confidence, which may lead to delayed intervention after errors have already propagated. We introduce Entropy-Trend Constraint (ETC), a training-free method that determines optimal retrieval timing by modeling the dynamics of token-level uncertainty. Specifically, ETC utilizes first- and second-order differences of the entropy sequence to detect emerging uncertainty trends, enabling earlier and more precise retrieval. Experiments on six QA benchmarks with three LLM backbones demonstrate that ETC consistently outperforms strong baselines while reducing retrieval frequency. ETC is particularly effective in domain-specific scenarios, exhibiting robust generalization capabilities. Ablation studies and qualitative analyses further confirm that trend-aware uncertainty modeling yields more effective retrieval timing. The method is plug-and-play, model-agnostic, and readily integrable into existing decoding pipelines. Implementation code is included in the supplementary materials.
MPL: Multiple Programming Languages with Large Language Models for Information Extraction
May 22, 2025Abstract:Recent research in information extraction (IE) focuses on utilizing code-style inputs to enhance structured output generation. The intuition behind this is that the programming languages (PLs) inherently exhibit greater structural organization than natural languages (NLs). This structural advantage makes PLs particularly suited for IE tasks. Nevertheless, existing research primarily focuses on Python for code-style simulation, overlooking the potential of other widely-used PLs (e.g., C++ and Java) during the supervised fine-tuning (SFT) phase. In this research, we propose \textbf{M}ultiple \textbf{P}rogramming \textbf{L}anguages with large language models for information extraction (abbreviated as \textbf{MPL}), a novel framework that explores the potential of incorporating different PLs in the SFT phase. Additionally, we introduce \texttt{function-prompt} with virtual running to simulate code-style inputs more effectively and efficiently. Experimental results on a wide range of datasets demonstrate the effectiveness of MPL. Furthermore, we conduct extensive experiments to provide a comprehensive analysis. We have released our code for future research.
C^2M-DoT: Cross-modal consistent multi-view medical report generation with domain transfer network
Oct 09, 2023Abstract:In clinical scenarios, multiple medical images with different views are usually generated simultaneously, and these images have high semantic consistency. However, most existing medical report generation methods only consider single-view data. The rich multi-view mutual information of medical images can help generate more accurate reports, however, the dependence of multi-view models on multi-view data in the inference stage severely limits their application in clinical practice. In addition, word-level optimization based on numbers ignores the semantics of reports and medical images, and the generated reports often cannot achieve good performance. Therefore, we propose a cross-modal consistent multi-view medical report generation with a domain transfer network (C^2M-DoT). Specifically, (i) a semantic-based multi-view contrastive learning medical report generation framework is adopted to utilize cross-view information to learn the semantic representation of lesions; (ii) a domain transfer network is further proposed to ensure that the multi-view report generation model can still achieve good inference performance under single-view input; (iii) meanwhile, optimization using a cross-modal consistency loss facilitates the generation of textual reports that are semantically consistent with medical images. Extensive experimental studies on two public benchmark datasets demonstrate that C^2M-DoT substantially outperforms state-of-the-art baselines in all metrics. Ablation studies also confirmed the validity and necessity of each component in C^2M-DoT.
AMLP:Adaptive Masking Lesion Patches for Self-supervised Medical Image Segmentation
Sep 08, 2023Abstract:Self-supervised masked image modeling has shown promising results on natural images. However, directly applying such methods to medical images remains challenging. This difficulty stems from the complexity and distinct characteristics of lesions compared to natural images, which impedes effective representation learning. Additionally, conventional high fixed masking ratios restrict reconstructing fine lesion details, limiting the scope of learnable information. To tackle these limitations, we propose a novel self-supervised medical image segmentation framework, Adaptive Masking Lesion Patches (AMLP). Specifically, we design a Masked Patch Selection (MPS) strategy to identify and focus learning on patches containing lesions. Lesion regions are scarce yet critical, making their precise reconstruction vital. To reduce misclassification of lesion and background patches caused by unsupervised clustering in MPS, we introduce an Attention Reconstruction Loss (ARL) to focus on hard-to-reconstruct patches likely depicting lesions. We further propose a Category Consistency Loss (CCL) to refine patch categorization based on reconstruction difficulty, strengthening distinction between lesions and background. Moreover, we develop an Adaptive Masking Ratio (AMR) strategy that gradually increases the masking ratio to expand reconstructible information and improve learning. Extensive experiments on two medical segmentation datasets demonstrate AMLP's superior performance compared to existing self-supervised approaches. The proposed strategies effectively address limitations in applying masked modeling to medical images, tailored to capturing fine lesion details vital for segmentation tasks.
MvCo-DoT:Multi-View Contrastive Domain Transfer Network for Medical Report Generation
Apr 15, 2023Abstract:In clinical scenarios, multiple medical images with different views are usually generated at the same time, and they have high semantic consistency. However, the existing medical report generation methods cannot exploit the rich multi-view mutual information of medical images. Therefore, in this work, we propose the first multi-view medical report generation model, called MvCo-DoT. Specifically, MvCo-DoT first propose a multi-view contrastive learning (MvCo) strategy to help the deep reinforcement learning based model utilize the consistency of multi-view inputs for better model learning. Then, to close the performance gaps of using multi-view and single-view inputs, a domain transfer network is further proposed to ensure MvCo-DoT achieve almost the same performance as multi-view inputs using only single-view inputs.Extensive experiments on the IU X-Ray public dataset show that MvCo-DoT outperforms the SOTA medical report generation baselines in all metrics.
MPS-AMS: Masked Patches Selection and Adaptive Masking Strategy Based Self-Supervised Medical Image Segmentation
Feb 27, 2023Abstract:Existing self-supervised learning methods based on contrastive learning and masked image modeling have demonstrated impressive performances. However, current masked image modeling methods are mainly utilized in natural images, and their applications in medical images are relatively lacking. Besides, their fixed high masking strategy limits the upper bound of conditional mutual information, and the gradient noise is considerable, making less the learned representation information. Motivated by these limitations, in this paper, we propose masked patches selection and adaptive masking strategy based self-supervised medical image segmentation method, named MPS-AMS. We leverage the masked patches selection strategy to choose masked patches with lesions to obtain more lesion representation information, and the adaptive masking strategy is utilized to help learn more mutual information and improve performance further. Extensive experiments on three public medical image segmentation datasets (BUSI, Hecktor, and Brats2018) show that our proposed method greatly outperforms the state-of-the-art self-supervised baselines.
Multi-Head Feature Pyramid Networks for Breast Mass Detection
Feb 22, 2023Abstract:Analysis of X-ray images is one of the main tools to diagnose breast cancer. The ability to quickly and accurately detect the location of masses from the huge amount of image data is the key to reducing the morbidity and mortality of breast cancer. Currently, the main factor limiting the accuracy of breast mass detection is the unequal focus on the mass boxes, leading the network to focus too much on larger masses at the expense of smaller ones. In the paper, we propose the multi-head feature pyramid module (MHFPN) to solve the problem of unbalanced focus of target boxes during feature map fusion and design a multi-head breast mass detection network (MBMDnet). Experimental studies show that, comparing to the SOTA detection baselines, our method improves by 6.58% (in AP@50) and 5.4% (in TPR@50) on the commonly used INbreast dataset, while about 6-8% improvements (in AP@20) are also observed on the public MIAS and BCS-DBT datasets.
Incremental Predictive Coding: A Parallel and Fully Automatic Learning Algorithm
Nov 16, 2022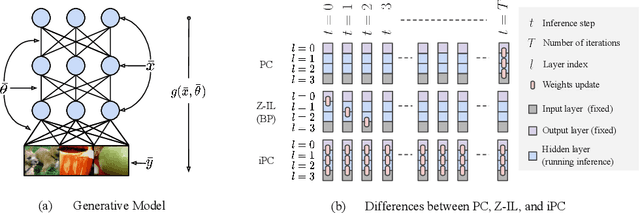
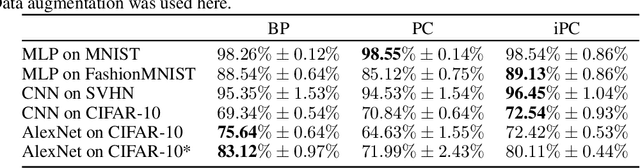
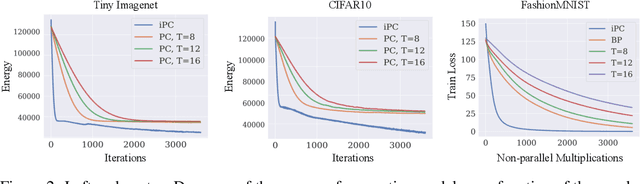

Abstract:Neuroscience-inspired models, such as predictive coding, have the potential to play an important role in the future of machine intelligence. However, they are not yet used in industrial applications due to some limitations, such as the lack of efficiency. In this work, we address this by proposing incremental predictive coding (iPC), a variation of the original framework derived from the incremental expectation maximization algorithm, where every operation can be performed in parallel without external control. We show both theoretically and empirically that iPC is much faster than the original algorithm originally developed by Rao and Ballard, while maintaining performance comparable to backpropagation in image classification tasks. This work impacts several areas, has general applications in computational neuroscience and machine learning, and specific applications in scenarios where automatization and parallelization are important, such as distributed computing and implementations of deep learning models on analog and neuromorphic chips.
PCA: Semi-supervised Segmentation with Patch Confidence Adversarial Training
Jul 24, 2022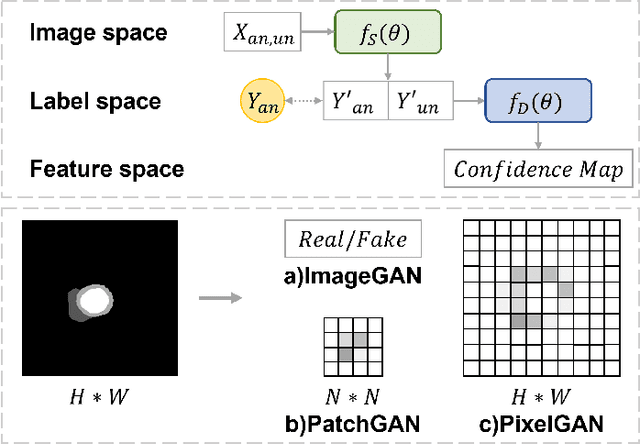
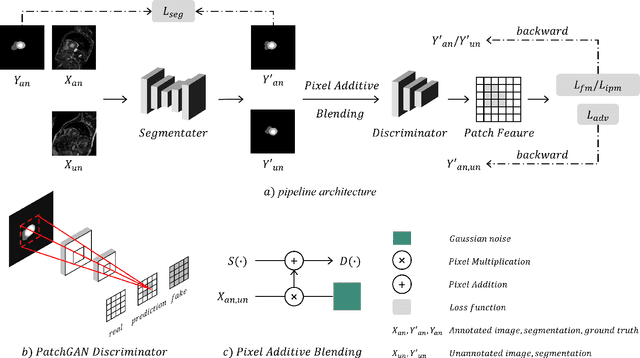
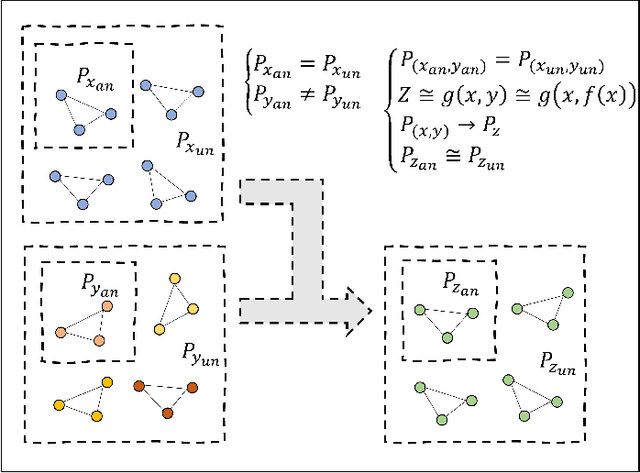
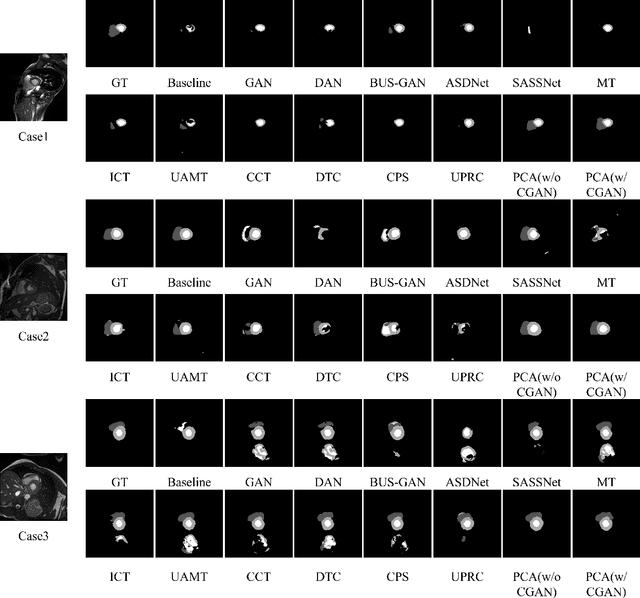
Abstract:Deep learning based semi-supervised learning (SSL) methods have achieved strong performance in medical image segmentation, which can alleviate doctors' expensive annotation by utilizing a large amount of unlabeled data. Unlike most existing semi-supervised learning methods, adversarial training based methods distinguish samples from different sources by learning the data distribution of the segmentation map, leading the segmenter to generate more accurate predictions. We argue that the current performance restrictions for such approaches are the problems of feature extraction and learning preference. In this paper, we propose a new semi-supervised adversarial method called Patch Confidence Adversarial Training (PCA) for medical image segmentation. Rather than single scalar classification results or pixel-level confidence maps, our proposed discriminator creates patch confidence maps and classifies them at the scale of the patches. The prediction of unlabeled data learns the pixel structure and context information in each patch to get enough gradient feedback, which aids the discriminator in convergent to an optimal state and improves semi-supervised segmentation performance. Furthermore, at the discriminator's input, we supplement semantic information constraints on images, making it simpler for unlabeled data to fit the expected data distribution. Extensive experiments on the Automated Cardiac Diagnosis Challenge (ACDC) 2017 dataset and the Brain Tumor Segmentation (BraTS) 2019 challenge dataset show that our method outperforms the state-of-the-art semi-supervised methods, which demonstrates its effectiveness for medical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge