Yohei Sugawara
Breaking the Data Barrier: Robust Few-Shot 3D Vessel Segmentation using Foundation Models
Feb 27, 2026Abstract:State-of-the-art vessel segmentation methods typically require large-scale annotated datasets and suffer from severe performance degradation under domain shifts. In clinical practice, however, acquiring extensive annotations for every new scanner or protocol is unfeasible. To address this, we propose a novel framework leveraging a pre-trained Vision Foundation Model (DINOv3) adapted for volumetric vessel segmentation. We introduce a lightweight 3D Adapter for volumetric consistency, a multi-scale 3D Aggregator for hierarchical feature fusion, and Z-channel embedding to effectively bridge the gap between 2D pre-training and 3D medical modalities, enabling the model to capture continuous vascular structures from limited data. We validated our method on the TopCoW (in-domain) and Lausanne (out-of-distribution) datasets. In the extreme few-shot regime with 5 training samples, our method achieved a Dice score of 43.42%, marking a 30% relative improvement over the state-of-the-art nnU-Net (33.41%) and outperforming other Transformer-based baselines, such as SwinUNETR and UNETR, by up to 45%. Furthermore, in the out-of-distribution setting, our model demonstrated superior robustness, achieving a 50% relative improvement over nnU-Net (21.37% vs. 14.22%), which suffered from severe domain overfitting. Ablation studies confirmed that our 3D adaptation mechanism and multi-scale aggregation strategy are critical for vascular continuity and robustness. Our results suggest foundation models offer a viable cold-start solution, improving clinical reliability under data scarcity or domain shifts.
Anatomical 3D Style Transfer Enabling Efficient Federated Learning with Extremely Low Communication Costs
Oct 26, 2024Abstract:In this study, we propose a novel federated learning (FL) approach that utilizes 3D style transfer for the multi-organ segmentation task. The multi-organ dataset, obtained by integrating multiple datasets, has high scalability and can improve generalization performance as the data volume increases. However, the heterogeneity of data owing to different clients with diverse imaging conditions and target organs can lead to severe overfitting of local models. To align models that overfit to different local datasets, existing methods require frequent communication with the central server, resulting in higher communication costs and risk of privacy leakage. To achieve an efficient and safe FL, we propose an Anatomical 3D Frequency Domain Generalization (A3DFDG) method for FL. A3DFDG utilizes structural information of human organs and clusters the 3D styles based on the location of organs. By mixing styles based on these clusters, it preserves the anatomical information and leads models to learn intra-organ diversity, while aligning the optimization of each local model. Experiments indicate that our method can maintain its accuracy even in cases where the communication cost is highly limited (=1.25% of the original cost) while achieving a significant difference compared to baselines, with a higher global dice similarity coefficient score of 4.3%. Despite its simplicity and minimal computational overhead, these results demonstrate that our method has high practicality in real-world scenarios where low communication costs and a simple pipeline are required. The code used in this project will be publicly available.
Virtual Human Generative Model: Masked Modeling Approach for Learning Human Characteristics
Jun 19, 2023



Abstract:Identifying the relationship between healthcare attributes, lifestyles, and personality is vital for understanding and improving physical and mental conditions. Machine learning approaches are promising for modeling their relationships and offering actionable suggestions. In this paper, we propose Virtual Human Generative Model (VHGM), a machine learning model for estimating attributes about healthcare, lifestyles, and personalities. VHGM is a deep generative model trained with masked modeling to learn the joint distribution of attributes conditioned on known ones. Using heterogeneous tabular datasets, VHGM learns more than 1,800 attributes efficiently. We numerically evaluate the performance of VHGM and its training techniques. As a proof-of-concept of VHGM, we present several applications demonstrating user scenarios, such as virtual measurements of healthcare attributes and hypothesis verifications of lifestyles.
Label-Efficient Multi-Task Segmentation using Contrastive Learning
Sep 23, 2020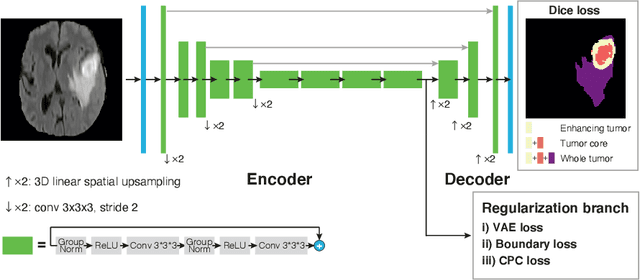
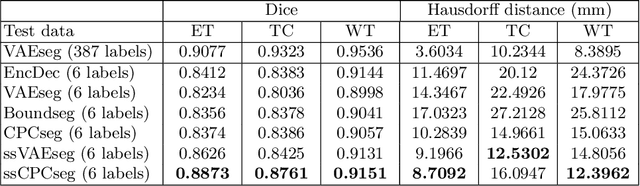
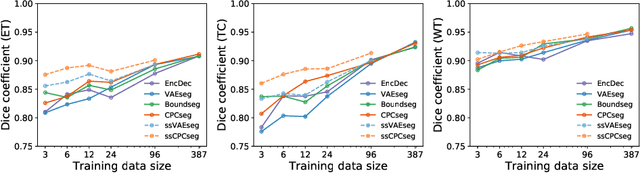
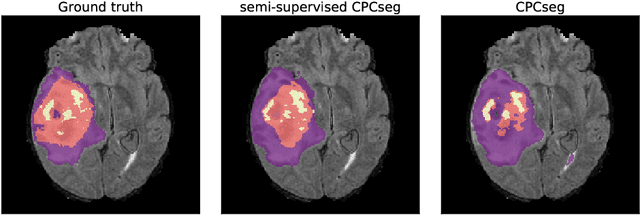
Abstract:Obtaining annotations for 3D medical images is expensive and time-consuming, despite its importance for automating segmentation tasks. Although multi-task learning is considered an effective method for training segmentation models using small amounts of annotated data, a systematic understanding of various subtasks is still lacking. In this study, we propose a multi-task segmentation model with a contrastive learning based subtask and compare its performance with other multi-task models, varying the number of labeled data for training. We further extend our model so that it can utilize unlabeled data through the regularization branch in a semi-supervised manner. We experimentally show that our proposed method outperforms other multi-task methods including the state-of-the-art fully supervised model when the amount of annotated data is limited.
An Inductive Transfer Learning Approach using Cycle-consistent Adversarial Domain Adaptation with Application to Brain Tumor Segmentation
May 11, 2020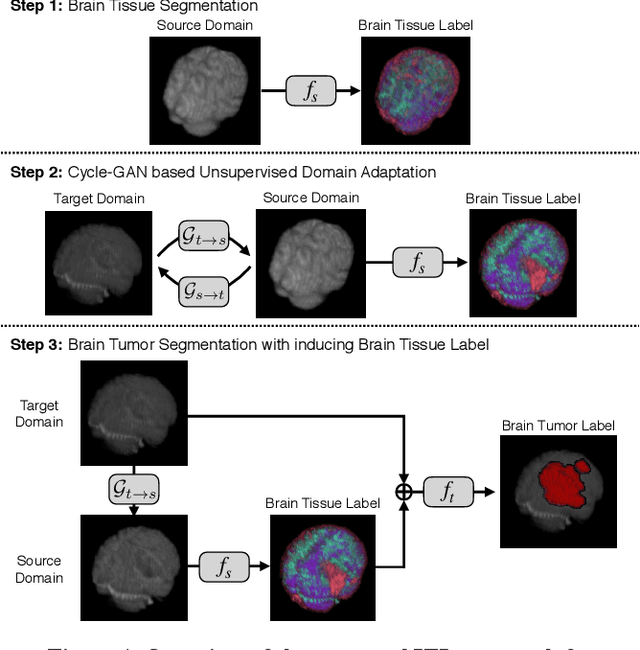
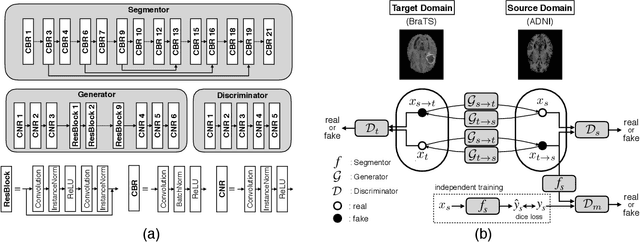
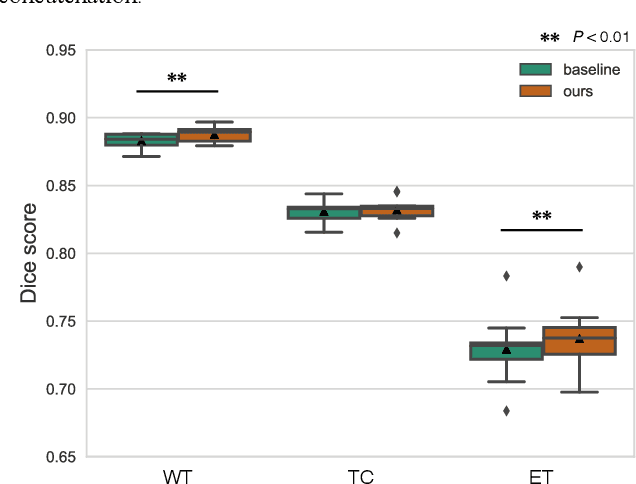
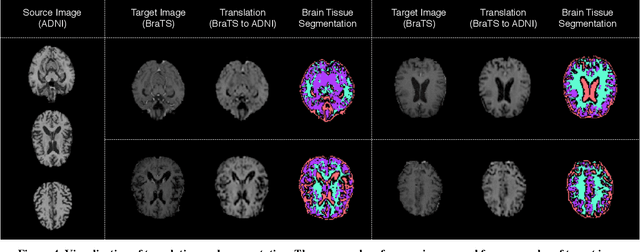
Abstract:With recent advances in supervised machine learning for medical image analysis applications, the annotated medical image datasets of various domains are being shared extensively. Given that the annotation labelling requires medical expertise, such labels should be applied to as many learning tasks as possible. However, the multi-modal nature of each annotated image renders it difficult to share the annotation label among diverse tasks. In this work, we provide an inductive transfer learning (ITL) approach to adopt the annotation label of the source domain datasets to tasks of the target domain datasets using Cycle-GAN based unsupervised domain adaptation (UDA). To evaluate the applicability of the ITL approach, we adopted the brain tissue annotation label on the source domain dataset of Magnetic Resonance Imaging (MRI) images to the task of brain tumor segmentation on the target domain dataset of MRI. The results confirm that the segmentation accuracy of brain tumor segmentation improved significantly. The proposed ITL approach can make significant contribution to the field of medical image analysis, as we develop a fundamental tool to improve and promote various tasks using medical images.
GA-GAN: CT reconstruction from Biplanar DRRs using GAN with Guided Attention
Oct 13, 2019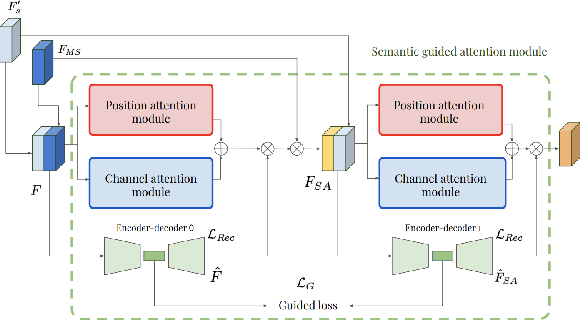
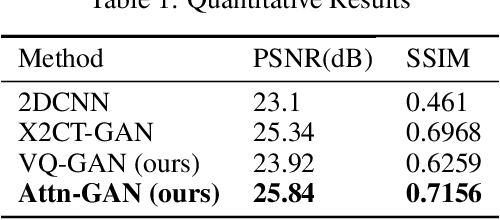
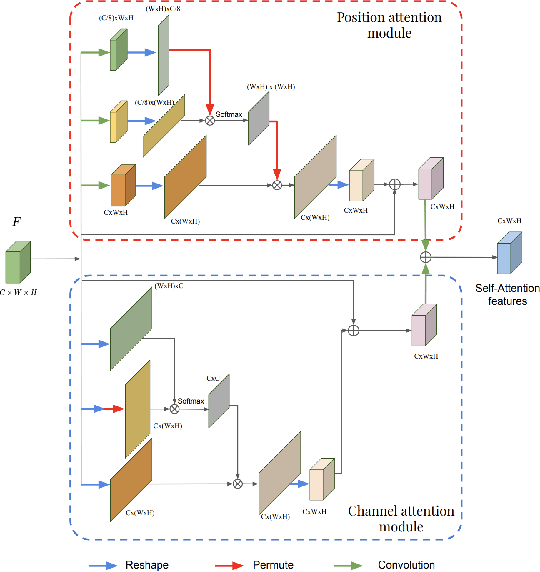
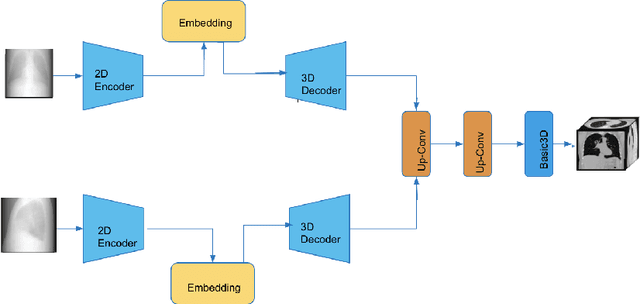
Abstract:This work investigates the use of guided attention in the reconstruction of CTvolumes from biplanar DRRs. We try to improve the visual image quality of the CT reconstruction using Guided Attention based GANs (GA-GAN). We also consider the use of Vector Quantization (VQ) for the CT reconstruction so that the memory usage can be reduced, maintaining the same visual image quality. To the best of our knowledge no work has been done before that explores the Vector Quantization for this purpose. Although our findings show that our approaches outperform the previous works, still there is a lot of room for improvement.
Einconv: Exploring Unexplored Tensor Decompositions for Convolutional Neural Networks
Aug 13, 2019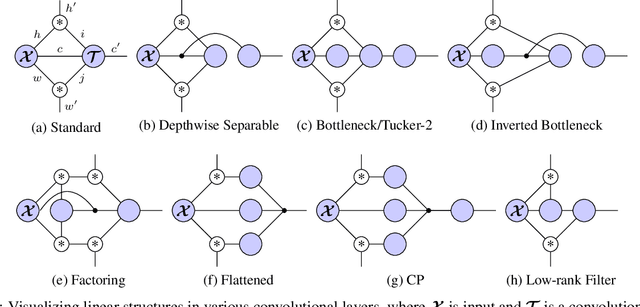
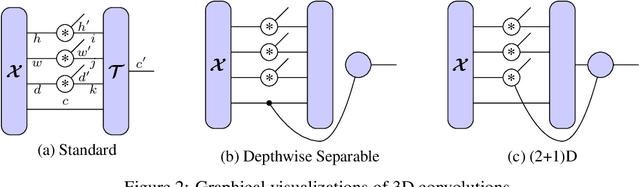
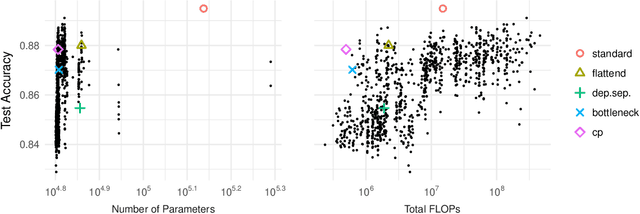
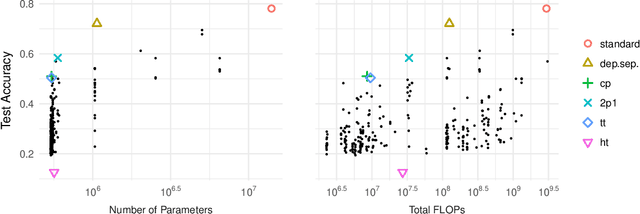
Abstract:Tensor decomposition methods are one of the primary approaches for model compression and fast inference of convolutional neural networks (CNNs). However, despite their potential diversity, only a few typical decompositions such as CP decomposition have been applied in practice; more importantly, no extensive comparisons have been performed between available methods. This raises the simple question of how many decompositions are possible, and which of these is the best. In this paper, we first characterize a decomposition class specific to CNNs by adopting graphical notation, which is considerably flexible. When combining with the nonlinear activations, the class includes renowned CNN modules such as depthwise separable convolution and bottleneck layer. In the experiments, we compare the tradeoff between prediction accuracy and time/space complexities by enumerating all the possible decompositions. Also, we demonstrate, using a neural architecture search, that we can find nonlinear decompositions that outperform existing decompositions.
BayesGrad: Explaining Predictions of Graph Convolutional Networks
Jul 04, 2018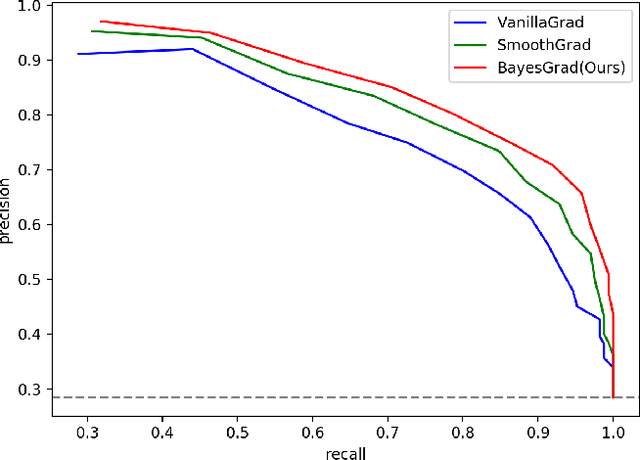
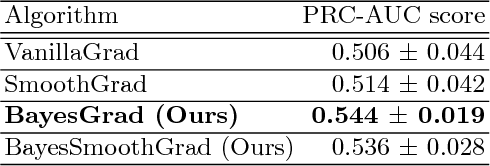
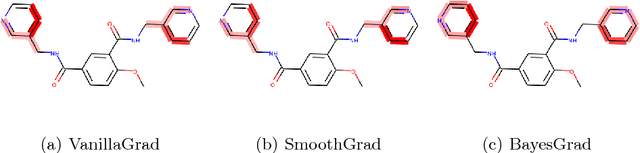
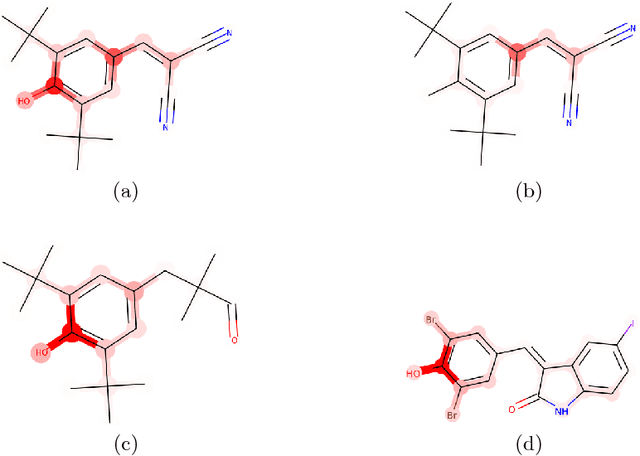
Abstract:Recent advances in graph convolutional networks have significantly improved the performance of chemical predictions, raising a new research question: "how do we explain the predictions of graph convolutional networks?" A possible approach to answer this question is to visualize evidence substructures responsible for the predictions. For chemical property prediction tasks, the sample size of the training data is often small and/or a label imbalance problem occurs, where a few samples belong to a single class and the majority of samples belong to the other classes. This can lead to uncertainty related to the learned parameters of the machine learning model. To address this uncertainty, we propose BayesGrad, utilizing the Bayesian predictive distribution, to define the importance of each node in an input graph, which is computed efficiently using the dropout technique. We demonstrate that BayesGrad successfully visualizes the substructures responsible for the label prediction in the artificial experiment, even when the sample size is small. Furthermore, we use a real dataset to evaluate the effectiveness of the visualization. The basic idea of BayesGrad is not limited to graph-structured data and can be applied to other data types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge