Yaoyun Zhang
A Study of Large Language Models for Patient Information Extraction: Model Architecture, Fine-Tuning Strategy, and Multi-task Instruction Tuning
Sep 05, 2025



Abstract:Natural language processing (NLP) is a key technology to extract important patient information from clinical narratives to support healthcare applications. The rapid development of large language models (LLMs) has revolutionized many NLP tasks in the clinical domain, yet their optimal use in patient information extraction tasks requires further exploration. This study examines LLMs' effectiveness in patient information extraction, focusing on LLM architectures, fine-tuning strategies, and multi-task instruction tuning techniques for developing robust and generalizable patient information extraction systems. This study aims to explore key concepts of using LLMs for clinical concept and relation extraction tasks, including: (1) encoder-only or decoder-only LLMs, (2) prompt-based parameter-efficient fine-tuning (PEFT) algorithms, and (3) multi-task instruction tuning on few-shot learning performance. We benchmarked a suite of LLMs, including encoder-based LLMs (BERT, GatorTron) and decoder-based LLMs (GatorTronGPT, Llama 3.1, GatorTronLlama), across five datasets. We compared traditional full-size fine-tuning and prompt-based PEFT. We explored a multi-task instruction tuning framework that combines both tasks across four datasets to evaluate the zero-shot and few-shot learning performance using the leave-one-dataset-out strategy.
Smooth-Foley: Creating Continuous Sound for Video-to-Audio Generation Under Semantic Guidance
Dec 24, 2024

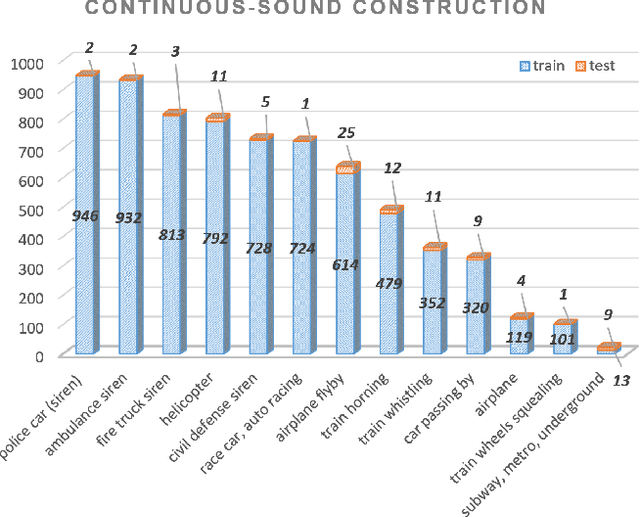
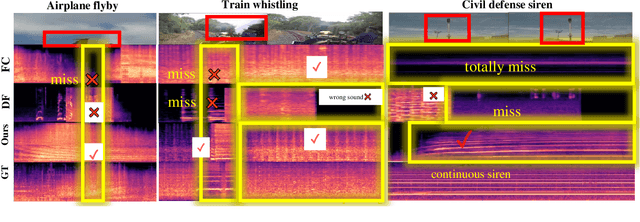
Abstract:The video-to-audio (V2A) generation task has drawn attention in the field of multimedia due to the practicality in producing Foley sound. Semantic and temporal conditions are fed to the generation model to indicate sound events and temporal occurrence. Recent studies on synthesizing immersive and synchronized audio are faced with challenges on videos with moving visual presence. The temporal condition is not accurate enough while low-resolution semantic condition exacerbates the problem. To tackle these challenges, we propose Smooth-Foley, a V2A generative model taking semantic guidance from the textual label across the generation to enhance both semantic and temporal alignment in audio. Two adapters are trained to leverage pre-trained text-to-audio generation models. A frame adapter integrates high-resolution frame-wise video features while a temporal adapter integrates temporal conditions obtained from similarities of visual frames and textual labels. The incorporation of semantic guidance from textual labels achieves precise audio-video alignment. We conduct extensive quantitative and qualitative experiments. Results show that Smooth-Foley performs better than existing models on both continuous sound scenarios and general scenarios. With semantic guidance, the audio generated by Smooth-Foley exhibits higher quality and better adherence to physical laws.
Domain Invariant Learning for Gaussian Processes and Bayesian Exploration
Dec 18, 2023Abstract:Out-of-distribution (OOD) generalization has long been a challenging problem that remains largely unsolved. Gaussian processes (GP), as popular probabilistic model classes, especially in the small data regime, presume strong OOD generalization abilities. Surprisingly, their OOD generalization abilities have been under-explored before compared with other lines of GP research. In this paper, we identify that GP is not free from the problem and propose a domain invariant learning algorithm for Gaussian processes (DIL-GP) with a min-max optimization on the likelihood. DIL-GP discovers the heterogeneity in the data and forces invariance across partitioned subsets of data. We further extend the DIL-GP to improve Bayesian optimization's adaptability on changing environments. Numerical experiments demonstrate the superiority of DIL-GP for predictions on several synthetic and real-world datasets. We further demonstrate the effectiveness of the DIL-GP Bayesian optimization method on a PID parameters tuning experiment for a quadrotor. The full version and source code are available at: https://github.com/Billzxl/DIL-GP.
DiffDTM: A conditional structure-free framework for bioactive molecules generation targeted for dual proteins
Jun 24, 2023Abstract:Advances in deep generative models shed light on de novo molecule generation with desired properties. However, molecule generation targeted for dual protein targets still faces formidable challenges including protein 3D structure data requisition for model training, auto-regressive sampling, and model generalization for unseen targets. Here, we proposed DiffDTM, a novel conditional structure-free deep generative model based on a diffusion model for dual targets based molecule generation to address the above issues. Specifically, DiffDTM receives protein sequences and molecular graphs as inputs instead of protein and molecular conformations and incorporates an information fusion module to achieve conditional generation in a one-shot manner. We have conducted comprehensive multi-view experiments to demonstrate that DiffDTM can generate drug-like, synthesis-accessible, novel, and high-binding affinity molecules targeting specific dual proteins, outperforming the state-of-the-art (SOTA) models in terms of multiple evaluation metrics. Furthermore, we utilized DiffDTM to generate molecules towards dopamine receptor D2 and 5-hydroxytryptamine receptor 1A as new antipsychotics. The experimental results indicate that DiffDTM can be easily plugged into unseen dual targets to generate bioactive molecules, addressing the issues of requiring insufficient active molecule data for training as well as the need to retrain when encountering new targets.
Revisiting Automatic Question Summarization Evaluation in the Biomedical Domain
Mar 18, 2023



Abstract:Automatic evaluation metrics have been facilitating the rapid development of automatic summarization methods by providing instant and fair assessments of the quality of summaries. Most metrics have been developed for the general domain, especially news and meeting notes, or other language-generation tasks. However, these metrics are applied to evaluate summarization systems in different domains, such as biomedical question summarization. To better understand whether commonly used evaluation metrics are capable of evaluating automatic summarization in the biomedical domain, we conduct human evaluations of summarization quality from four different aspects of a biomedical question summarization task. Based on human judgments, we identify different noteworthy features for current automatic metrics and summarization systems as well. We also release a dataset of our human annotations to aid the research of summarization evaluation metrics in the biomedical domain.
Molecular Geometry-aware Transformer for accurate 3D Atomic System modeling
Feb 02, 2023



Abstract:Molecular dynamic simulations are important in computational physics, chemistry, material, and biology. Machine learning-based methods have shown strong abilities in predicting molecular energy and properties and are much faster than DFT calculations. Molecular energy is at least related to atoms, bonds, bond angles, torsion angles, and nonbonding atom pairs. Previous Transformer models only use atoms as inputs which lack explicit modeling of the aforementioned factors. To alleviate this limitation, we propose Moleformer, a novel Transformer architecture that takes nodes (atoms) and edges (bonds and nonbonding atom pairs) as inputs and models the interactions among them using rotational and translational invariant geometry-aware spatial encoding. Proposed spatial encoding calculates relative position information including distances and angles among nodes and edges. We benchmark Moleformer on OC20 and QM9 datasets, and our model achieves state-of-the-art on the initial state to relaxed energy prediction of OC20 and is very competitive in QM9 on predicting quantum chemical properties compared to other Transformer and Graph Neural Network (GNN) methods which proves the effectiveness of the proposed geometry-aware spatial encoding in Moleformer.
COVID-19 SignSym: A fast adaptation of general clinical NLP tools to identify and normalize COVID-19 signs and symptoms to OMOP common data model
Jul 13, 2020



Abstract:The COVID-19 pandemic swept across the world rapidly infecting millions of people. An efficient tool that can accurately recognize important clinical concepts of COVID-19 from free text in electronic health records will be significantly valuable to accelerate various applications of COVID-19 research. To this end, the existing clinical NLP tool CLAMP was quickly adapted to COVID-19 information and generated an automated tool called COVID-19 SignSym, which can extract and signs/symptoms and their eight attributes such as temporal information and negations from clinical text. The extracted information is also mapped to standard clinical concepts in the common data model of OHDSI OMOP. Evaluation on clinical notes and medical dialogues demonstrated promising results. It is freely accessible to the community as a downloadable package of APIs (https://clamp.uth.edu/covid/nlp.php). We believe COVID-19 SignSym will provide fundamental supports to the secondary use of EHRs, thus accelerating the global research of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge