Masoud Rouhizadeh
N3C Natural Language Processing
An Open Natural Language Processing Development Framework for EHR-based Clinical Research: A case demonstration using the National COVID Cohort Collaborative (N3C)
Oct 20, 2021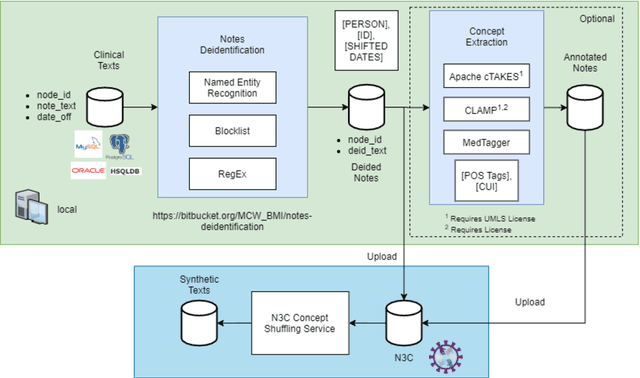
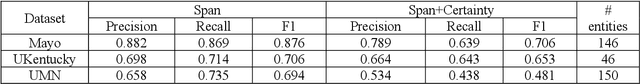
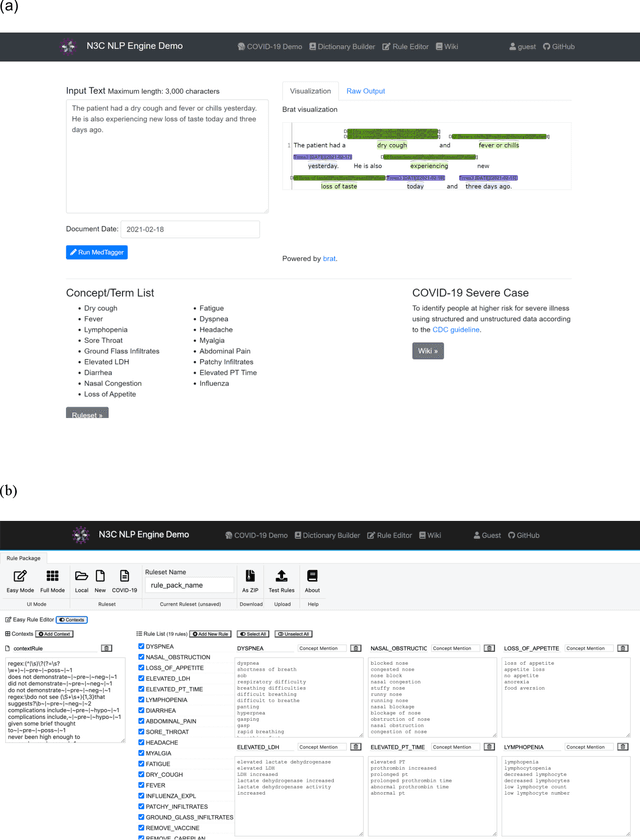
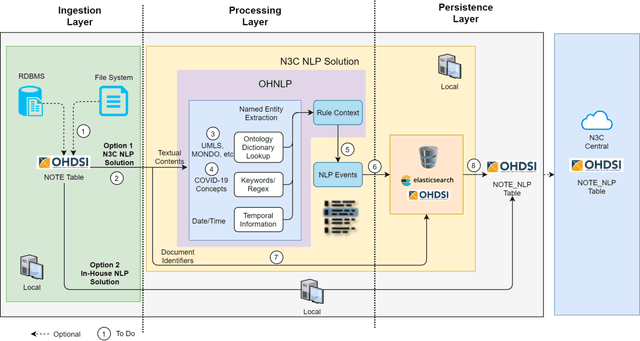
Abstract:While we pay attention to the latest advances in clinical natural language processing (NLP), we can notice some resistance in the clinical and translational research community to adopt NLP models due to limited transparency, Interpretability and usability. Built upon our previous work, in this study, we proposed an open natural language processing development framework and evaluated it through the implementation of NLP algorithms for the National COVID Cohort Collaborative (N3C). Based on the interests in information extraction from COVID-19 related clinical notes, our work includes 1) an open data annotation process using COVID-19 signs and symptoms as the use case, 2) a community-driven ruleset composing platform, and 3) a synthetic text data generation workflow to generate texts for information extraction tasks without involving human subjects. The generated corpora derived out of the texts from multiple intuitions and gold standard annotation are tested on a single institution's rule set has the performances in F1 score of 0.876, 0.706 and 0.694, respectively. The study as a consortium effort of the N3C NLP subgroup demonstrates the feasibility of creating a federated NLP algorithm development and benchmarking platform to enhance multi-institution clinical NLP study.
COVID-19 SignSym: A fast adaptation of general clinical NLP tools to identify and normalize COVID-19 signs and symptoms to OMOP common data model
Jul 13, 2020



Abstract:The COVID-19 pandemic swept across the world rapidly infecting millions of people. An efficient tool that can accurately recognize important clinical concepts of COVID-19 from free text in electronic health records will be significantly valuable to accelerate various applications of COVID-19 research. To this end, the existing clinical NLP tool CLAMP was quickly adapted to COVID-19 information and generated an automated tool called COVID-19 SignSym, which can extract and signs/symptoms and their eight attributes such as temporal information and negations from clinical text. The extracted information is also mapped to standard clinical concepts in the common data model of OHDSI OMOP. Evaluation on clinical notes and medical dialogues demonstrated promising results. It is freely accessible to the community as a downloadable package of APIs (https://clamp.uth.edu/covid/nlp.php). We believe COVID-19 SignSym will provide fundamental supports to the secondary use of EHRs, thus accelerating the global research of COVID-19.
Phenotyping of Clinical Notes with Improved Document Classification Models Using Contextualized Neural Language Models
Oct 30, 2019

Abstract:Clinical notes contain an extensive record of a patient's health status, such as smoking status or the presence of heart conditions. However, this detail is not replicated within the structured data of electronic health systems. Phenotyping, the extraction of patient conditions from free clinical text, is a critical task which supports avariety of downstream applications such as decision support and secondary use ofmedical records. Previous work has resulted in systems which are high performing but require hand engineering, often of rules. Recent work in pretrained contextualized language models have enabled advances in representing text for a variety of tasks. We therefore explore several architectures for modeling pheno-typing that rely solely on BERT representations of the clinical note, removing the need for manual engineering. We find these architectures are competitive with or outperform existing state of the art methods on two phenotyping tasks.
Deriving Verb Predicates By Clustering Verbs with Arguments
Aug 01, 2017



Abstract:Hand-built verb clusters such as the widely used Levin classes (Levin, 1993) have proved useful, but have limited coverage. Verb classes automatically induced from corpus data such as those from VerbKB (Wijaya, 2016), on the other hand, can give clusters with much larger coverage, and can be adapted to specific corpora such as Twitter. We present a method for clustering the outputs of VerbKB: verbs with their multiple argument types, e.g. "marry(person, person)", "feel(person, emotion)." We make use of a novel low-dimensional embedding of verbs and their arguments to produce high quality clusters in which the same verb can be in different clusters depending on its argument type. The resulting verb clusters do a better job than hand-built clusters of predicting sarcasm, sentiment, and locus of control in tweets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge