Virginia Adams
RedPajama: an Open Dataset for Training Large Language Models
Nov 19, 2024Abstract:Large language models are increasingly becoming a cornerstone technology in artificial intelligence, the sciences, and society as a whole, yet the optimal strategies for dataset composition and filtering remain largely elusive. Many of the top-performing models lack transparency in their dataset curation and model development processes, posing an obstacle to the development of fully open language models. In this paper, we identify three core data-related challenges that must be addressed to advance open-source language models. These include (1) transparency in model development, including the data curation process, (2) access to large quantities of high-quality data, and (3) availability of artifacts and metadata for dataset curation and analysis. To address these challenges, we release RedPajama-V1, an open reproduction of the LLaMA training dataset. In addition, we release RedPajama-V2, a massive web-only dataset consisting of raw, unfiltered text data together with quality signals and metadata. Together, the RedPajama datasets comprise over 100 trillion tokens spanning multiple domains and with their quality signals facilitate the filtering of data, aiming to inspire the development of numerous new datasets. To date, these datasets have already been used in the training of strong language models used in production, such as Snowflake Arctic, Salesforce's XGen and AI2's OLMo. To provide insight into the quality of RedPajama, we present a series of analyses and ablation studies with decoder-only language models with up to 1.6B parameters. Our findings demonstrate how quality signals for web data can be effectively leveraged to curate high-quality subsets of the dataset, underscoring the potential of RedPajama to advance the development of transparent and high-performing language models at scale.
HelpSteer: Multi-attribute Helpfulness Dataset for SteerLM
Nov 16, 2023

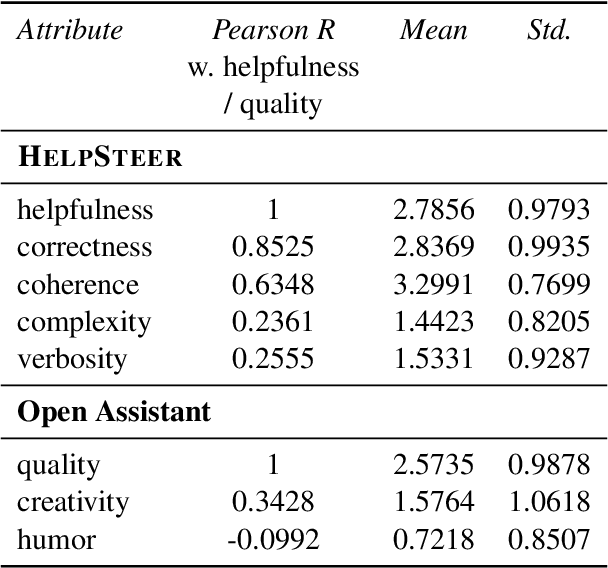
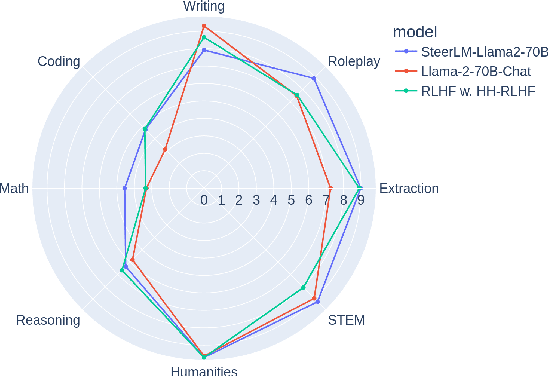
Abstract:Existing open-source helpfulness preference datasets do not specify what makes some responses more helpful and others less so. Models trained on these datasets can incidentally learn to model dataset artifacts (e.g. preferring longer but unhelpful responses only due to their length). To alleviate this problem, we collect HelpSteer, a multi-attribute helpfulness dataset annotated for the various aspects that make responses helpful. Specifically, our 37k-sample dataset has annotations for correctness, coherence, complexity, and verbosity in addition to overall helpfulness of responses. Training Llama 2 70B using the HelpSteer dataset with SteerLM technique produces a model that scores 7.54 on MT Bench, which is currently the highest score for open models that do not require training data from more powerful models (e.g. GPT4). We release this dataset with CC-BY-4.0 license at https://huggingface.co/datasets/nvidia/HelpSteer
Evaluating Parameter Efficient Learning for Generation
Oct 25, 2022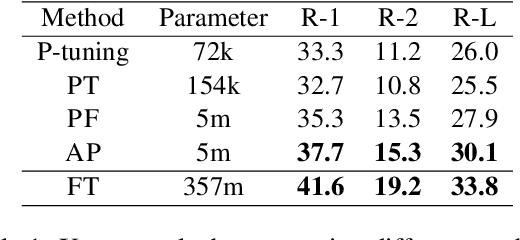
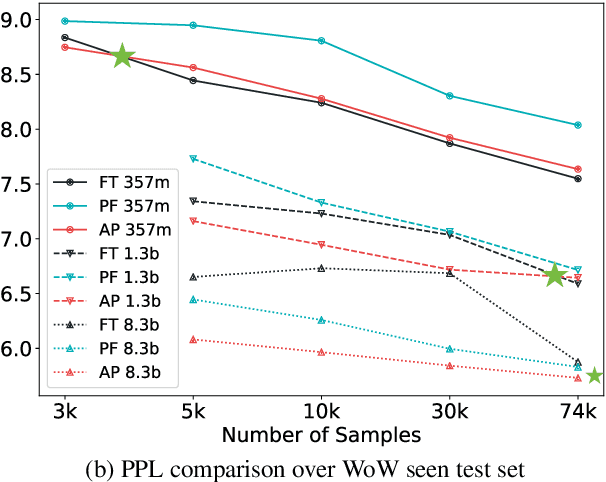
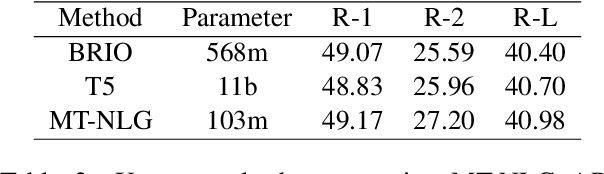
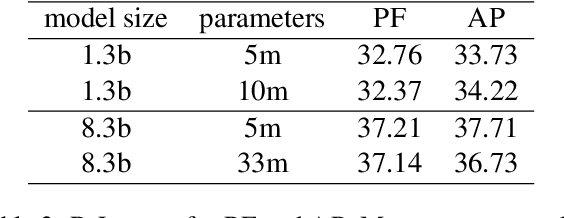
Abstract:Parameter efficient learning methods (PERMs) have recently gained significant attention as they provide an efficient way for pre-trained language models (PLMs) to adapt to a downstream task. However, these conclusions are mostly drawn from in-domain evaluations over the full training set. In this paper, we present comparisons between PERMs and finetuning from three new perspectives: (1) the effect of sample and model size to in-domain evaluations, (2) generalization to unseen domains and new datasets, and (3) the faithfulness of generations. Our results show that for in-domain settings (a) there is a cross point of sample size for which PERMs will perform better than finetuning when training with fewer samples, and (b) larger PLMs have larger cross points. For cross-domain and cross-dataset cases, we show that (a) Adapter (Houlsby et al., 2019) performs the best amongst all the PERMs studied here, and (b) it outperforms finetuning if the task dataset is below a certain size. We also compare the faithfulness of generations and show that PERMs can achieve better faithfulness score than finetuning, especially for small training set, by as much as 6%. Finally, we apply Adapter to MT-NLG 530b (Smith et al., 2022) and achieve new state-of-the-art results on Xsum (Narayan et al., 2018) for all ROUGE scores (ROUGE-1 49.17, ROUGE-2 27.20, ROUGE-L 40.98).
Finding the Right Recipe for Low Resource Domain Adaptation in Neural Machine Translation
Jun 02, 2022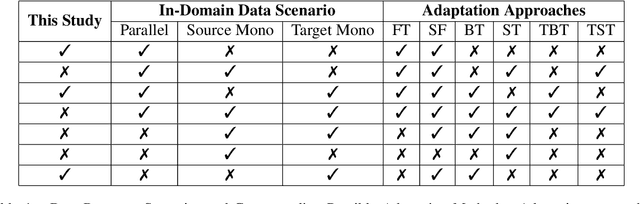
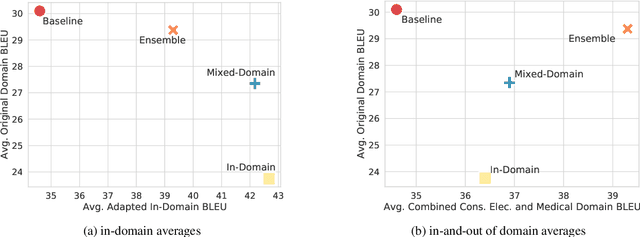
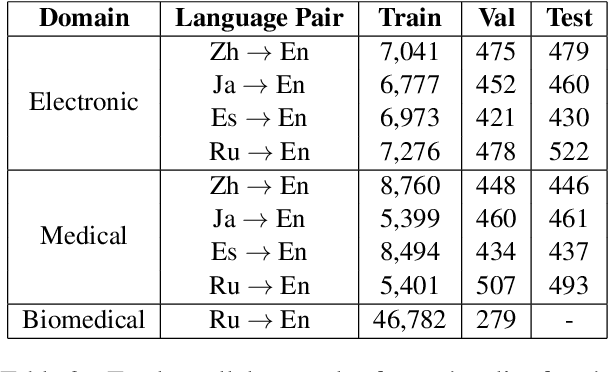
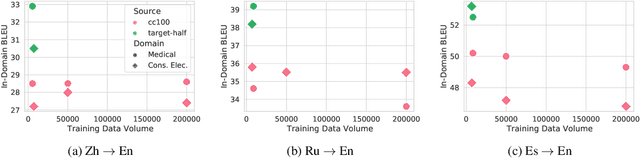
Abstract:General translation models often still struggle to generate accurate translations in specialized domains. To guide machine translation practitioners and characterize the effectiveness of domain adaptation methods under different data availability scenarios, we conduct an in-depth empirical exploration of monolingual and parallel data approaches to domain adaptation of pre-trained, third-party, NMT models in settings where architecture change is impractical. We compare data centric adaptation methods in isolation and combination. We study method effectiveness in very low resource (8k parallel examples) and moderately low resource (46k parallel examples) conditions and propose an ensemble approach to alleviate reductions in original domain translation quality. Our work includes three domains: consumer electronic, clinical, and biomedical and spans four language pairs - Zh-En, Ja-En, Es-En, and Ru-En. We also make concrete recommendations for achieving high in-domain performance and release our consumer electronic and medical domain datasets for all languages and make our code publicly available.
Automatic Extraction of Medication Names in Tweets as Named Entity Recognition
Nov 30, 2021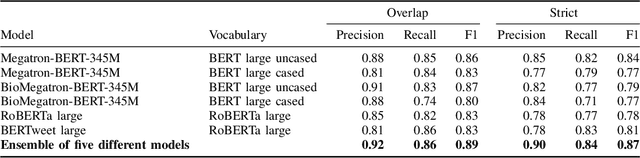
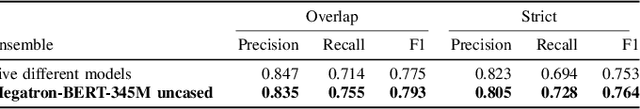
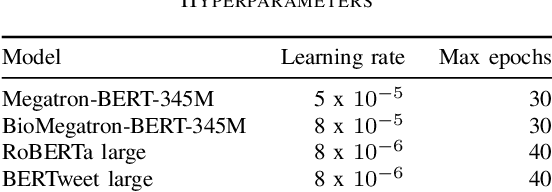
Abstract:Social media posts contain potentially valuable information about medical conditions and health-related behavior. Biocreative VII Task 3 focuses on mining this information by recognizing mentions of medications and dietary supplements in tweets. We approach this task by fine tuning multiple BERT-style language models to perform token-level classification, and combining them into ensembles to generate final predictions. Our best system consists of five Megatron-BERT-345M models and achieves a strict F1 score of 0.764 on unseen test data.
Chemical Identification and Indexing in PubMed Articles via BERT and Text-to-Text Approaches
Nov 30, 2021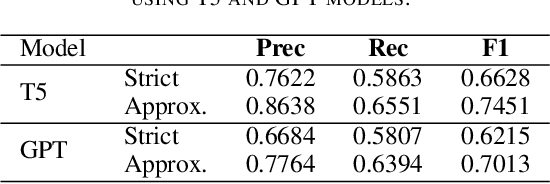

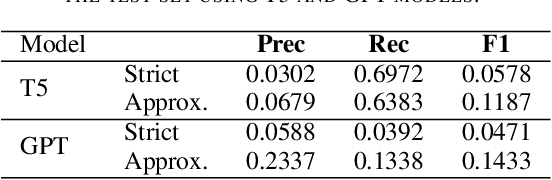
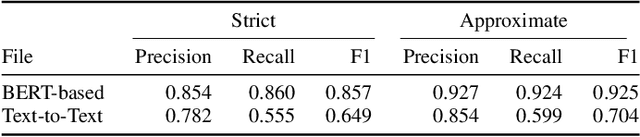
Abstract:The Biocreative VII Track-2 challenge consists of named entity recognition, entity-linking (or entity-normalization), and topic indexing tasks -- with entities and topics limited to chemicals for this challenge. Named entity recognition is a well-established problem and we achieve our best performance with BERT-based BioMegatron models. We extend our BERT-based approach to the entity linking task. After the second stage of pretraining BioBERT with a metric-learning loss strategy called self-alignment pretraining (SAP), we link entities based on the cosine similarity between their SAP-BioBERT word embeddings. Despite the success of our named entity recognition experiments, we find the chemical indexing task generally more challenging. In addition to conventional NER methods, we attempt both named entity recognition and entity linking with a novel text-to-text or "prompt" based method that uses generative language models such as T5 and GPT. We achieve encouraging results with this new approach.
Text Mining Drug/Chemical-Protein Interactions using an Ensemble of BERT and T5 Based Models
Nov 30, 2021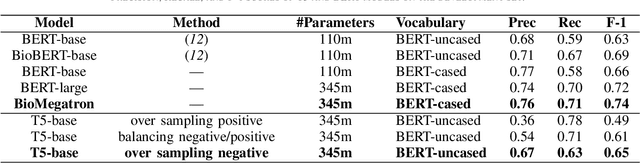

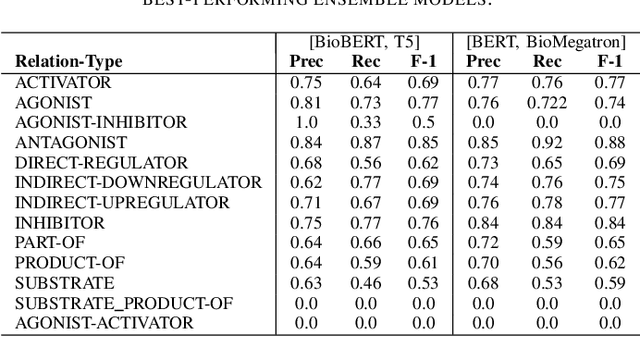
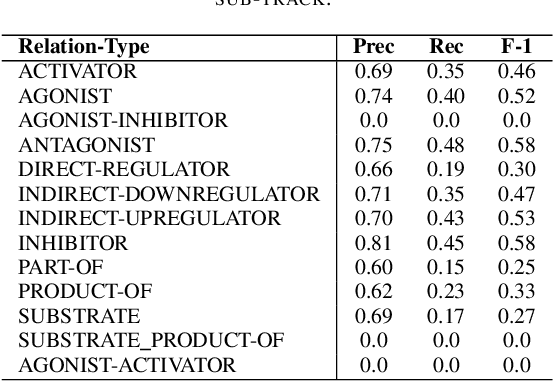
Abstract:In Track-1 of the BioCreative VII Challenge participants are asked to identify interactions between drugs/chemicals and proteins. In-context named entity annotations for each drug/chemical and protein are provided and one of fourteen different interactions must be automatically predicted. For this relation extraction task, we attempt both a BERT-based sentence classification approach, and a more novel text-to-text approach using a T5 model. We find that larger BERT-based models perform better in general, with our BioMegatron-based model achieving the highest scores across all metrics, achieving 0.74 F1 score. Though our novel T5 text-to-text method did not perform as well as most of our BERT-based models, it outperformed those trained on similar data, showing promising results, achieving 0.65 F1 score. We believe a text-to-text approach to relation extraction has some competitive advantages and there is a lot of room for research advancement.
NVIDIA NeMo Neural Machine Translation Systems for English-German and English-Russian News and Biomedical Tasks at WMT21
Nov 16, 2021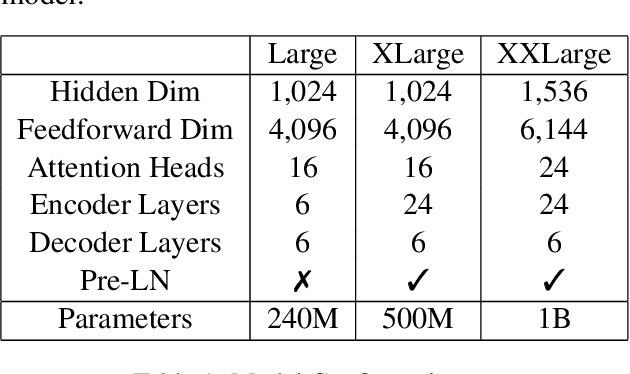
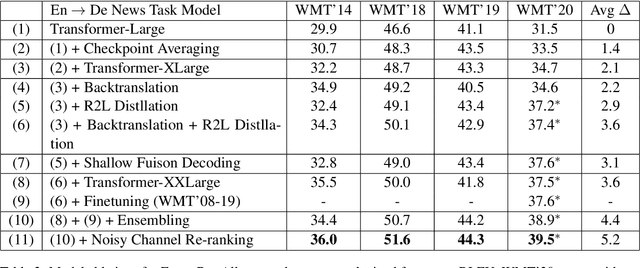
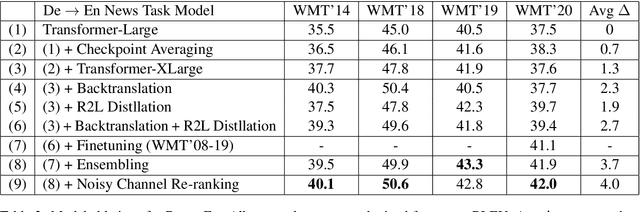
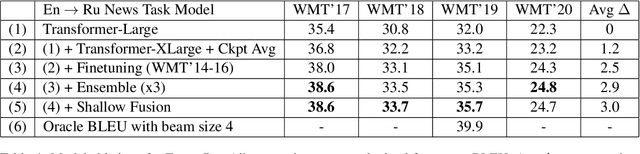
Abstract:This paper provides an overview of NVIDIA NeMo's neural machine translation systems for the constrained data track of the WMT21 News and Biomedical Shared Translation Tasks. Our news task submissions for English-German (En-De) and English-Russian (En-Ru) are built on top of a baseline transformer-based sequence-to-sequence model. Specifically, we use a combination of 1) checkpoint averaging 2) model scaling 3) data augmentation with backtranslation and knowledge distillation from right-to-left factorized models 4) finetuning on test sets from previous years 5) model ensembling 6) shallow fusion decoding with transformer language models and 7) noisy channel re-ranking. Additionally, our biomedical task submission for English-Russian uses a biomedically biased vocabulary and is trained from scratch on news task data, medically relevant text curated from the news task dataset, and biomedical data provided by the shared task. Our news system achieves a sacreBLEU score of 39.5 on the WMT'20 En-De test set outperforming the best submission from last year's task of 38.8. Our biomedical task Ru-En and En-Ru systems reach BLEU scores of 43.8 and 40.3 respectively on the WMT'20 Biomedical Task Test set, outperforming the previous year's best submissions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge