Carol Anderson
Ancestry.com
Consistency in Language Models: Current Landscape, Challenges, and Future Directions
May 01, 2025Abstract:The hallmark of effective language use lies in consistency -- expressing similar meanings in similar contexts and avoiding contradictions. While human communication naturally demonstrates this principle, state-of-the-art language models struggle to maintain reliable consistency across different scenarios. This paper examines the landscape of consistency research in AI language systems, exploring both formal consistency (including logical rule adherence) and informal consistency (such as moral and factual coherence). We analyze current approaches to measure aspects of consistency, identify critical research gaps in standardization of definitions, multilingual assessment, and methods to improve consistency. Our findings point to an urgent need for robust benchmarks to measure and interdisciplinary approaches to ensure consistency in the application of language models on domain-specific tasks while preserving the utility and adaptability.
Segmenting Messy Text: Detecting Boundaries in Text Derived from Historical Newspaper Images
Dec 20, 2023

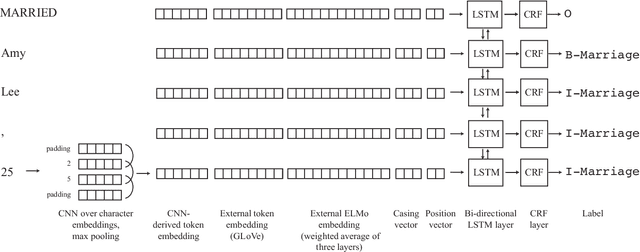

Abstract:Text segmentation, the task of dividing a document into sections, is often a prerequisite for performing additional natural language processing tasks. Existing text segmentation methods have typically been developed and tested using clean, narrative-style text with segments containing distinct topics. Here we consider a challenging text segmentation task: dividing newspaper marriage announcement lists into units of one announcement each. In many cases the information is not structured into sentences, and adjacent segments are not topically distinct from each other. In addition, the text of the announcements, which is derived from images of historical newspapers via optical character recognition, contains many typographical errors. As a result, these announcements are not amenable to segmentation with existing techniques. We present a novel deep learning-based model for segmenting such text and show that it significantly outperforms an existing state-of-the-art method on our task.
* 8 pages, 4 figures
Automatic Extraction of Medication Names in Tweets as Named Entity Recognition
Nov 30, 2021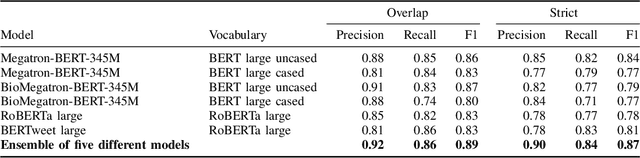
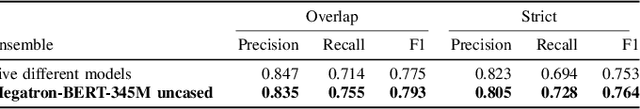
Abstract:Social media posts contain potentially valuable information about medical conditions and health-related behavior. Biocreative VII Task 3 focuses on mining this information by recognizing mentions of medications and dietary supplements in tweets. We approach this task by fine tuning multiple BERT-style language models to perform token-level classification, and combining them into ensembles to generate final predictions. Our best system consists of five Megatron-BERT-345M models and achieves a strict F1 score of 0.764 on unseen test data.
Chemical Identification and Indexing in PubMed Articles via BERT and Text-to-Text Approaches
Nov 30, 2021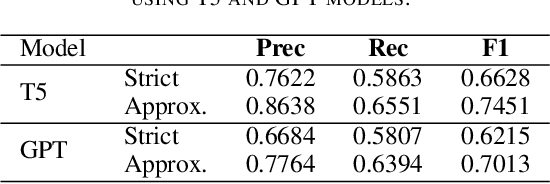

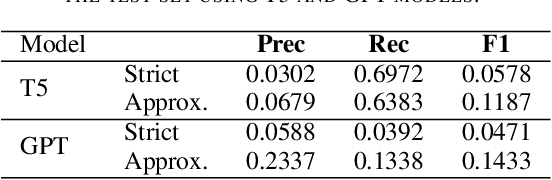
Abstract:The Biocreative VII Track-2 challenge consists of named entity recognition, entity-linking (or entity-normalization), and topic indexing tasks -- with entities and topics limited to chemicals for this challenge. Named entity recognition is a well-established problem and we achieve our best performance with BERT-based BioMegatron models. We extend our BERT-based approach to the entity linking task. After the second stage of pretraining BioBERT with a metric-learning loss strategy called self-alignment pretraining (SAP), we link entities based on the cosine similarity between their SAP-BioBERT word embeddings. Despite the success of our named entity recognition experiments, we find the chemical indexing task generally more challenging. In addition to conventional NER methods, we attempt both named entity recognition and entity linking with a novel text-to-text or "prompt" based method that uses generative language models such as T5 and GPT. We achieve encouraging results with this new approach.
Text Mining Drug/Chemical-Protein Interactions using an Ensemble of BERT and T5 Based Models
Nov 30, 2021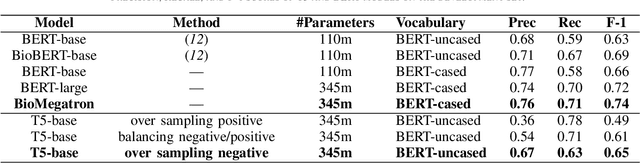

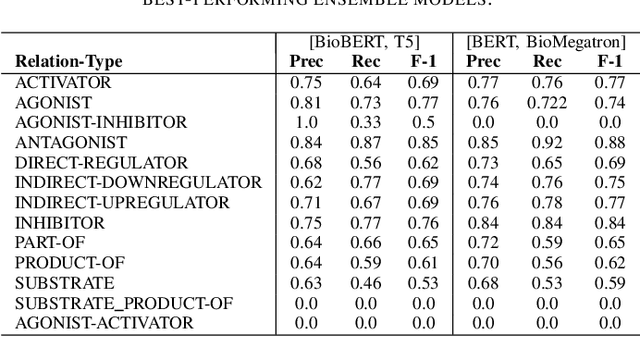
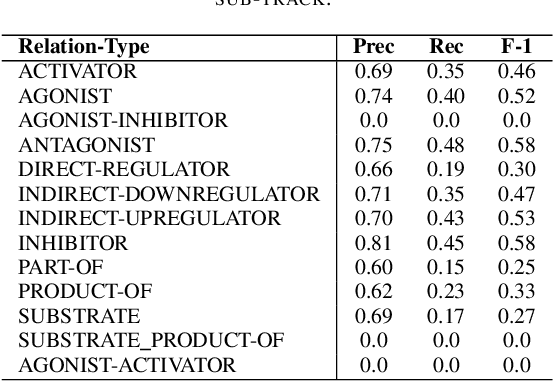
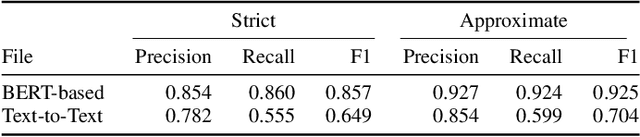
Abstract:In Track-1 of the BioCreative VII Challenge participants are asked to identify interactions between drugs/chemicals and proteins. In-context named entity annotations for each drug/chemical and protein are provided and one of fourteen different interactions must be automatically predicted. For this relation extraction task, we attempt both a BERT-based sentence classification approach, and a more novel text-to-text approach using a T5 model. We find that larger BERT-based models perform better in general, with our BioMegatron-based model achieving the highest scores across all metrics, achieving 0.74 F1 score. Though our novel T5 text-to-text method did not perform as well as most of our BERT-based models, it outperformed those trained on similar data, showing promising results, achieving 0.65 F1 score. We believe a text-to-text approach to relation extraction has some competitive advantages and there is a lot of room for research advancement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge