Anas Abidin
Investigation of large-scale extended Granger causality (lsXGC) on synthetic functional MRI data
May 06, 2022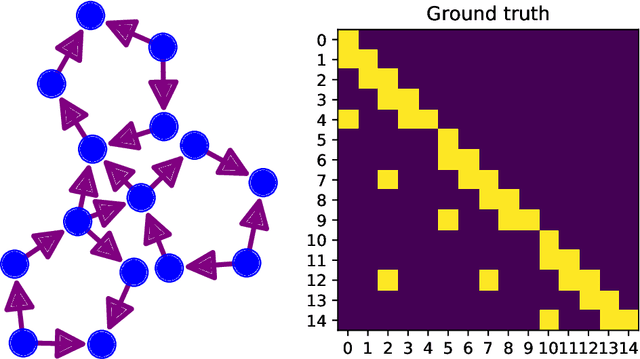
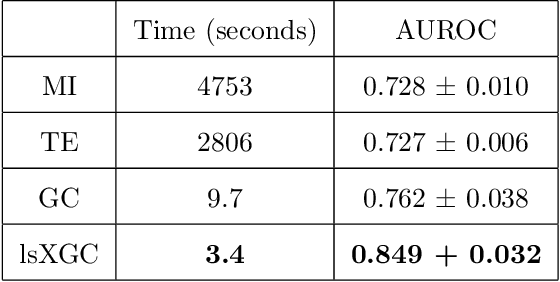
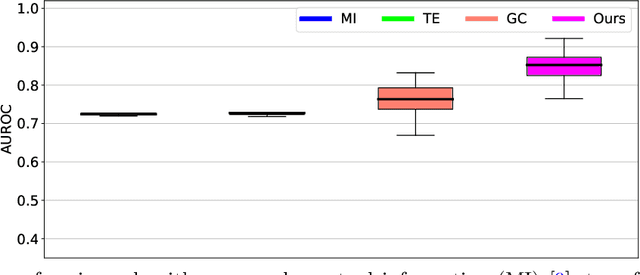
Abstract:It is a challenging research endeavor to infer causal relationships in multivariate observational time-series. Such data may be represented by graphs, where nodes represent time-series, and edges directed causal influence scores between them. If the number of nodes exceeds the number of temporal observations, conventional methods, such as standard Granger causality, are of limited value, because estimating free parameters of time-series predictors lead to underdetermined problems. A typical example for this situation is functional Magnetic Resonance Imaging (fMRI), where the number of nodal observations is large, usually ranging from $10^2$ to $10^5$ time-series, while the number of temporal observations is low, usually less than $10^3$. Hence, innovative approaches are required to address the challenges arising from such data sets. Recently, we have proposed the large-scale Extended Granger Causality (lsXGC) algorithm, which is based on augmenting a dimensionality-reduced representation of the system's state-space by supplementing data from the conditional source time-series taken from the original input space. Here, we apply lsXGC on synthetic fMRI data with known ground truth and compare its performance to state-of-the-art methods by leveraging the benefits of information-theoretic approaches. Our results suggest that the proposed lsXGC method significantly outperforms existing methods, both in diagnostic accuracy with Area Under the Receiver Operating Characteristic (AUROC = $0.849$ vs.~$[0.727, 0.762]$ for competing methods, $p<\!10^{-8}$), and computation time ($3.4$ sec vs.~[$9.7$, $4.8 \times 10^3$] sec for competing methods) benchmarks, demonstrating the potential of lsXGC for analyzing large-scale networks in neuroimaging studies of the human brain.
Automatic Extraction of Medication Names in Tweets as Named Entity Recognition
Nov 30, 2021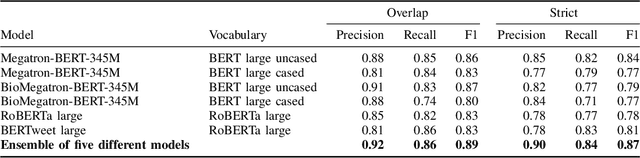
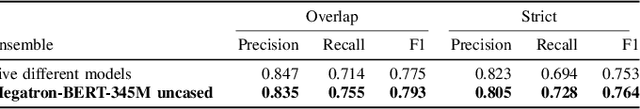
Abstract:Social media posts contain potentially valuable information about medical conditions and health-related behavior. Biocreative VII Task 3 focuses on mining this information by recognizing mentions of medications and dietary supplements in tweets. We approach this task by fine tuning multiple BERT-style language models to perform token-level classification, and combining them into ensembles to generate final predictions. Our best system consists of five Megatron-BERT-345M models and achieves a strict F1 score of 0.764 on unseen test data.
Chemical Identification and Indexing in PubMed Articles via BERT and Text-to-Text Approaches
Nov 30, 2021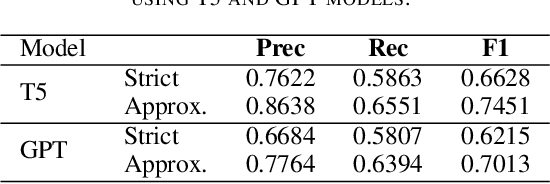

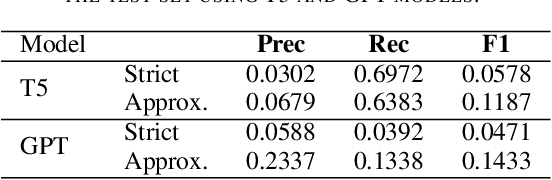
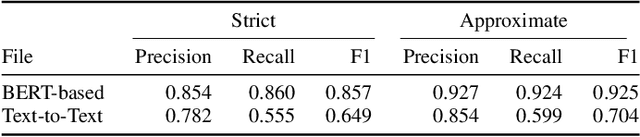
Abstract:The Biocreative VII Track-2 challenge consists of named entity recognition, entity-linking (or entity-normalization), and topic indexing tasks -- with entities and topics limited to chemicals for this challenge. Named entity recognition is a well-established problem and we achieve our best performance with BERT-based BioMegatron models. We extend our BERT-based approach to the entity linking task. After the second stage of pretraining BioBERT with a metric-learning loss strategy called self-alignment pretraining (SAP), we link entities based on the cosine similarity between their SAP-BioBERT word embeddings. Despite the success of our named entity recognition experiments, we find the chemical indexing task generally more challenging. In addition to conventional NER methods, we attempt both named entity recognition and entity linking with a novel text-to-text or "prompt" based method that uses generative language models such as T5 and GPT. We achieve encouraging results with this new approach.
Text Mining Drug/Chemical-Protein Interactions using an Ensemble of BERT and T5 Based Models
Nov 30, 2021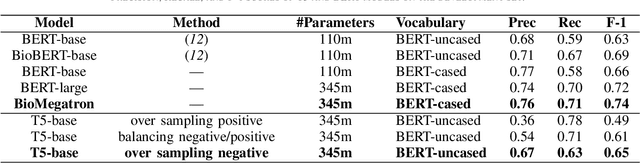

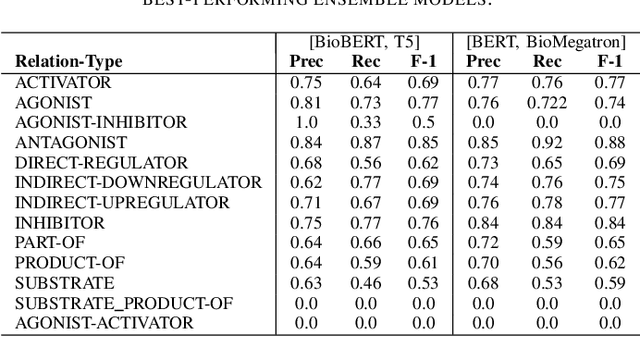
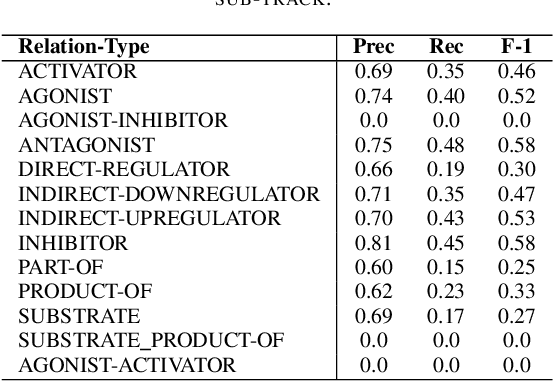
Abstract:In Track-1 of the BioCreative VII Challenge participants are asked to identify interactions between drugs/chemicals and proteins. In-context named entity annotations for each drug/chemical and protein are provided and one of fourteen different interactions must be automatically predicted. For this relation extraction task, we attempt both a BERT-based sentence classification approach, and a more novel text-to-text approach using a T5 model. We find that larger BERT-based models perform better in general, with our BioMegatron-based model achieving the highest scores across all metrics, achieving 0.74 F1 score. Though our novel T5 text-to-text method did not perform as well as most of our BERT-based models, it outperformed those trained on similar data, showing promising results, achieving 0.65 F1 score. We believe a text-to-text approach to relation extraction has some competitive advantages and there is a lot of room for research advancement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge