Axel Wismüller
Investigation of large-scale extended Granger causality (lsXGC) on synthetic functional MRI data
May 06, 2022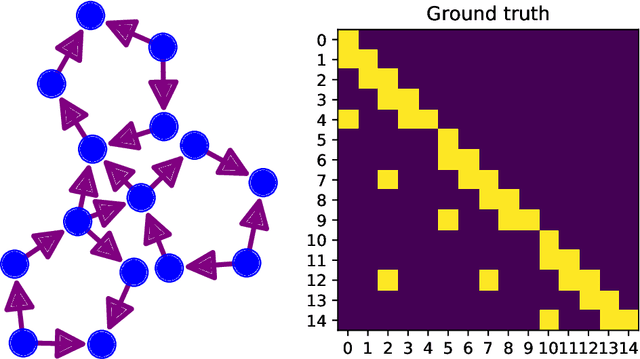
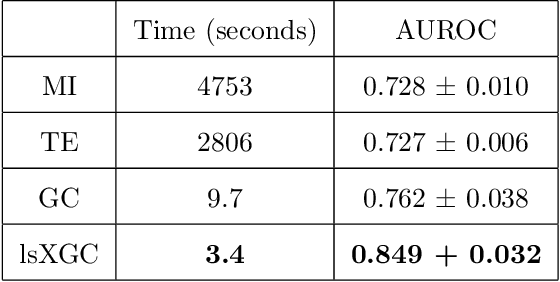
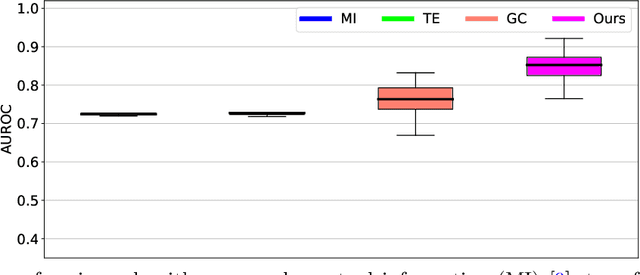
Abstract:It is a challenging research endeavor to infer causal relationships in multivariate observational time-series. Such data may be represented by graphs, where nodes represent time-series, and edges directed causal influence scores between them. If the number of nodes exceeds the number of temporal observations, conventional methods, such as standard Granger causality, are of limited value, because estimating free parameters of time-series predictors lead to underdetermined problems. A typical example for this situation is functional Magnetic Resonance Imaging (fMRI), where the number of nodal observations is large, usually ranging from $10^2$ to $10^5$ time-series, while the number of temporal observations is low, usually less than $10^3$. Hence, innovative approaches are required to address the challenges arising from such data sets. Recently, we have proposed the large-scale Extended Granger Causality (lsXGC) algorithm, which is based on augmenting a dimensionality-reduced representation of the system's state-space by supplementing data from the conditional source time-series taken from the original input space. Here, we apply lsXGC on synthetic fMRI data with known ground truth and compare its performance to state-of-the-art methods by leveraging the benefits of information-theoretic approaches. Our results suggest that the proposed lsXGC method significantly outperforms existing methods, both in diagnostic accuracy with Area Under the Receiver Operating Characteristic (AUROC = $0.849$ vs.~$[0.727, 0.762]$ for competing methods, $p<\!10^{-8}$), and computation time ($3.4$ sec vs.~[$9.7$, $4.8 \times 10^3$] sec for competing methods) benchmarks, demonstrating the potential of lsXGC for analyzing large-scale networks in neuroimaging studies of the human brain.
Classification of Schizophrenia from Functional MRI Using Large-scale Extended Granger Causality
Jan 12, 2021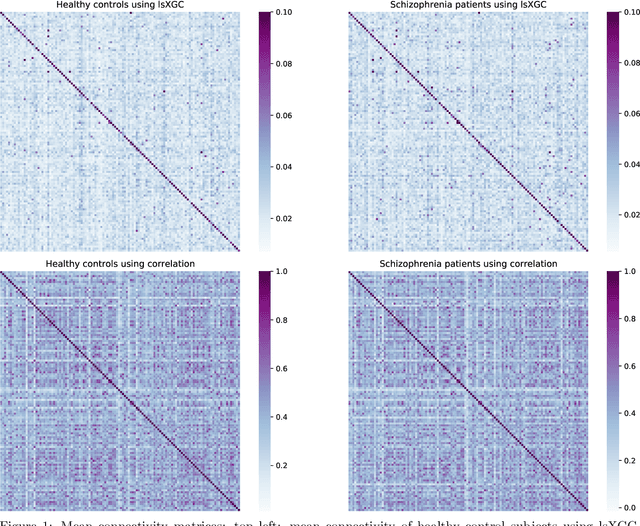
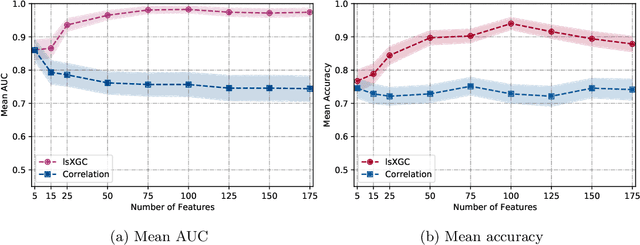
Abstract:The literature manifests that schizophrenia is associated with alterations in brain network connectivity. We investigate whether large-scale Extended Granger Causality (lsXGC) can capture such alterations using resting-state fMRI data. Our method utilizes dimension reduction combined with the augmentation of source time-series in a predictive time-series model for estimating directed causal relationships among fMRI time-series. The lsXGC is a multivariate approach since it identifies the relationship of the underlying dynamic system in the presence of all other time-series. Here lsXGC serves as a biomarker for classifying schizophrenia patients from typical controls using a subset of 62 subjects from the Centers of Biomedical Research Excellence (COBRE) data repository. We use brain connections estimated by lsXGC as features for classification. After feature extraction, we perform feature selection by Kendall's tau rank correlation coefficient followed by classification using a support vector machine. As a reference method, we compare our results with cross-correlation, typically used in the literature as a standard measure of functional connectivity. We cross-validate 100 different training/test (90%/10%) data split to obtain mean accuracy and a mean Area Under the receiver operating characteristic Curve (AUC) across all tested numbers of features for lsXGC. Our results demonstrate a mean accuracy range of [0.767, 0.940] and a mean AUC range of [0.861, 0.983] for lsXGC. The result of lsXGC is significantly higher than the results obtained with the cross-correlation, namely mean accuracy of [0.721, 0.751] and mean AUC of [0.744, 0.860]. Our results suggest the applicability of lsXGC as a potential biomarker for schizophrenia.
Tracking Results and Utilization of Artificial Intelligence (tru-AI) in Radiology: Early-Stage COVID-19 Pandemic Observations
Oct 14, 2020

Abstract:Objective: To introduce a method for tracking results and utilization of Artificial Intelligence (tru-AI) in radiology. By tracking both large-scale utilization and AI results data, the tru-AI approach is designed to calculate surrogates for measuring important disease-related observational quantities over time, such as the prevalence of intracranial hemorrhage during the COVID-19 pandemic outbreak. Methods: To quantitatively investigate the clinical applicability of the tru-AI approach, we analyzed service requests for automatically identifying intracranial hemorrhage (ICH) on head CT using a commercial AI solution. This software is typically used for AI-based prioritization of radiologists' reading lists for reducing turnaround times in patients with emergent clinical findings, such as ICH or pulmonary embolism.We analyzed data of N=9,421 emergency-setting non-contrast head CT studies at a major US healthcare system acquired from November 1, 2019 through June 2, 2020, and compared two observation periods, namely (i) a pre-pandemic epoch from November 1, 2019 through February 29, 2020, and (ii) a period during the COVID-19 pandemic outbreak, April 1-30, 2020. Results: Although daily CT scan counts were significantly lower during (40.1 +/- 7.9) than before (44.4 +/- 7.6) the COVID-19 outbreak, we found that ICH was more likely to be observed by AI during than before the COVID-19 outbreak (p<0.05), with approximately one daily ICH+ case more than statistically expected. Conclusion: Our results suggest that, by tracking both large-scale utilization and AI results data in radiology, the tru-AI approach can contribute clinical value as a versatile exploratory tool, aiming at a better understanding of pandemic-related effects on healthcare.
Large-scale nonlinear Granger causality: A data-driven, multivariate approach to recovering directed networks from short time-series data
Sep 10, 2020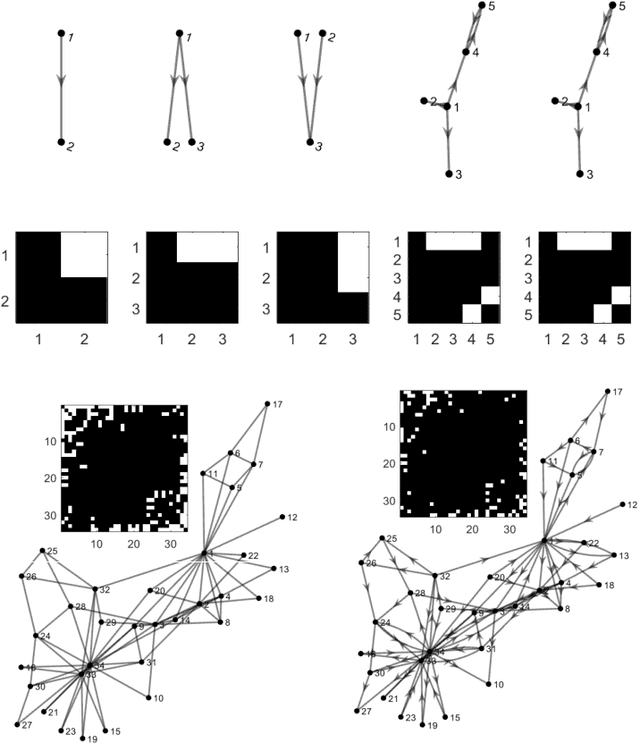
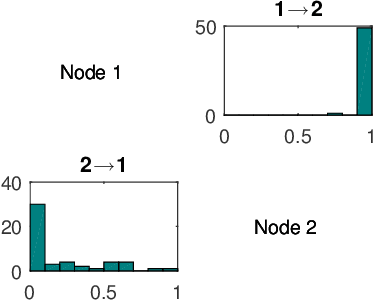
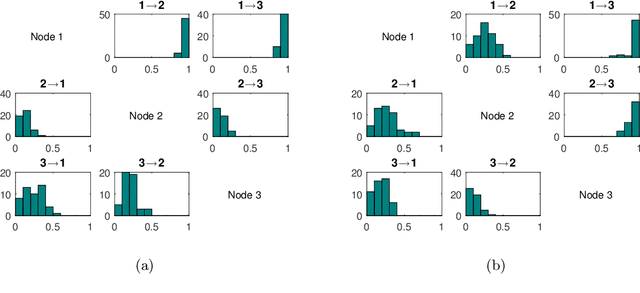
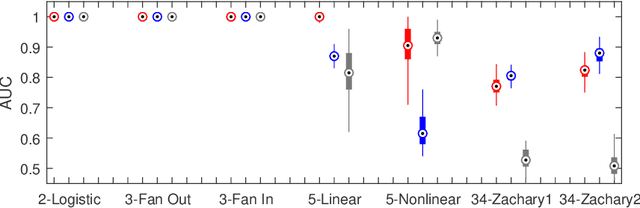
Abstract:To gain insight into complex systems it is a key challenge to infer nonlinear causal directional relations from observational time-series data. Specifically, estimating causal relationships between interacting components in large systems with only short recordings over few temporal observations remains an important, yet unresolved problem. Here, we introduce a large-scale Nonlinear Granger Causality (lsNGC) approach for inferring directional, nonlinear, multivariate causal interactions between system components from short high-dimensional time-series recordings. By modeling interactions with nonlinear state-space transformations from limited observational data, lsNGC identifies casual relations with no explicit a priori assumptions on functional interdependence between component time-series in a computationally efficient manner. Additionally, our method provides a mathematical formulation revealing statistical significance of inferred causal relations. We extensively study the ability of lsNGC to recovering network structure from two-node to thirty-four node chaotic time-series systems. Our results suggest that lsNGC captures meaningful interactions from limited observational data, where it performs favorably when compared to traditionally used methods. Finally, we demonstrate the applicability of lsNGC to estimating causality in large, real-world systems by inferring directional nonlinear, multivariate causal relationships among a large number of relatively short time-series acquired from functional Magnetic Resonance Imaging (fMRI) data of the human brain.
Automated Identification of Thoracic Pathology from Chest Radiographs with Enhanced Training Pipeline
Jun 11, 2020Abstract:Chest x-rays are the most common radiology studies for diagnosing lung and heart disease. Hence, a system for automated pre-reporting of pathologic findings on chest x-rays would greatly enhance radiologists' productivity. To this end, we investigate a deep-learning framework with novel training schemes for classification of different thoracic pathology labels from chest x-rays. We use the currently largest publicly available annotated dataset ChestX-ray14 of 112,120 chest radiographs of 30,805 patients. Each image was annotated with either a 'NoFinding' class, or one or more of 14 thoracic pathology labels. Subjects can have multiple pathologies, resulting in a multi-class, multi-label problem. We encoded labels as binary vectors using k-hot encoding. We study the ResNet34 architecture, pre-trained on ImageNet, where two key modifications were incorporated into the training framework: (1) Stochastic gradient descent with momentum and with restarts using cosine annealing, (2) Variable image sizes for fine-tuning to prevent overfitting. Additionally, we use a heuristic algorithm to select a good learning rate. Learning with restarts was used to avoid local minima. Area Under receiver operating characteristics Curve (AUC) was used to quantitatively evaluate diagnostic quality. Our results are comparable to, or outperform the best results of current state-of-the-art methods with AUCs as follows: Atelectasis:0.81, Cardiomegaly:0.91, Consolidation:0.81, Edema:0.92, Effusion:0.89, Emphysema: 0.92, Fibrosis:0.81, Hernia:0.84, Infiltration:0.73, Mass:0.85, Nodule:0.76, Pleural Thickening:0.81, Pneumonia:0.77, Pneumothorax:0.89 and NoFinding:0.79. Our results suggest that, in addition to using sophisticated network architectures, a good learning rate, scheduler and a robust optimizer can boost performance.
* 6 pages, 1 figure, 2 tables
A Framework for Exploring Non-Linear Functional Connectivity and Causality in the Human Brain: Mutual Connectivity Analysis (MCA) of Resting-State Functional MRI with Convergent Cross-Mapping and Non-Metric Clustering
Jul 14, 2014

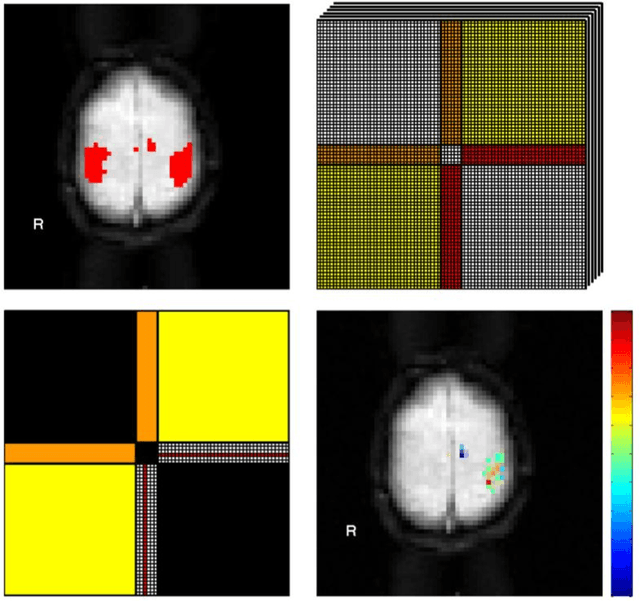

Abstract:We present a computational framework for analysis and visualization of non-linear functional connectivity in the human brain from resting state functional MRI (fMRI) data for purposes of recovering the underlying network community structure and exploring causality between network components. Our proposed methodology of non-linear mutual connectivity analysis (MCA) involves two computational steps. First, the pair-wise cross-prediction performance between resting state fMRI pixel time series within the brain is evaluated. The underlying network structure is subsequently recovered from the affinity matrix constructed through MCA using non-metric network partitioning/clustering with the so-called Louvain method. We demonstrate our methodology in the task of identifying regions of the motor cortex associated with hand movement on resting state fMRI data acquired from eight slice locations in four subjects. For comparison, we also localized regions of the motor cortex through a task-based fMRI sequence involving a finger-tapping stimulus paradigm. Finally, we integrate convergent cross mapping (CCM) into the first step of MCA for investigating causality between regions of the motor cortex. Results regarding causation between regions of the motor cortex revealed a significant directional variability and were not readily interpretable in a consistent manner across all subjects. However, our results on whole-slice fMRI analysis demonstrate that MCA-based model-free recovery of regions associated with the primary motor cortex and supplementary motor area are in close agreement with localization of similar regions achieved with a task-based fMRI acquisition. Thus, we conclude that our computational framework MCA can extract and visualize valuable information concerning the underlying network structure and causation between different regions of the brain in resting state fMRI.
A Computational Framework for Nonlinear Dimensionality Reduction of Large Data Sets: The Exploratory Inspection Machine (XIM)
Jun 10, 2011


Abstract:In this paper, we present a novel computational framework for nonlinear dimensionality reduction which is specifically suited to process large data sets: the Exploratory Inspection Machine (XIM). XIM introduces a conceptual cross-link between hitherto separate domains of machine learning, namely topographic vector quantization and divergence-based neighbor embedding approaches. There are three ways to conceptualize XIM, namely (i) as the inversion of the Exploratory Observation Machine (XOM) and its variants, such as Neighbor Embedding XOM (NE-XOM), (ii) as a powerful optimization scheme for divergence-based neighbor embedding cost functions inspired by Stochastic Neighbor Embedding (SNE) and its variants, such as t-distributed SNE (t-SNE), and (iii) as an extension of topographic vector quantization methods, such as the Self-Organizing Map (SOM). By preserving both global and local data structure, XIM combines the virtues of classical and advanced recent embedding methods. It permits direct visualization of large data collections without the need for prior data reduction. Finally, XIM can contribute to many application domains of data analysis and visualization important throughout the sciences and engineering, such as pattern matching, constrained incremental learning, data clustering, and the analysis of non-metric dissimilarity data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge