Classification of Schizophrenia from Functional MRI Using Large-scale Extended Granger Causality
Paper and Code
Jan 12, 2021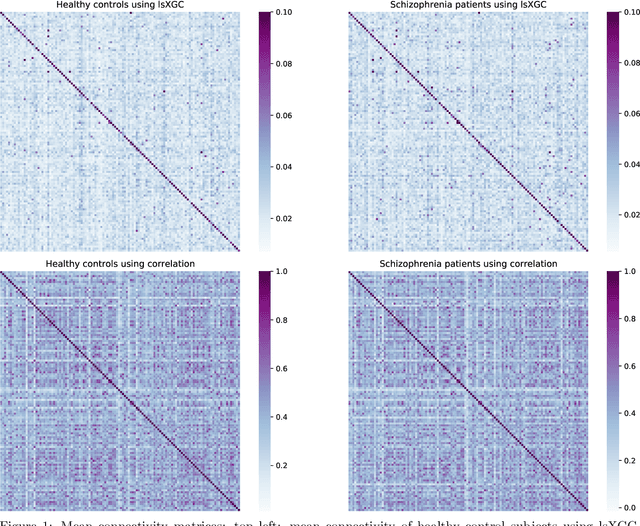
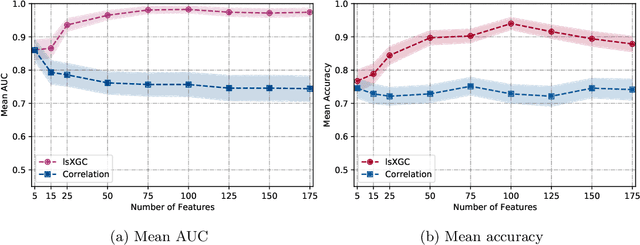
The literature manifests that schizophrenia is associated with alterations in brain network connectivity. We investigate whether large-scale Extended Granger Causality (lsXGC) can capture such alterations using resting-state fMRI data. Our method utilizes dimension reduction combined with the augmentation of source time-series in a predictive time-series model for estimating directed causal relationships among fMRI time-series. The lsXGC is a multivariate approach since it identifies the relationship of the underlying dynamic system in the presence of all other time-series. Here lsXGC serves as a biomarker for classifying schizophrenia patients from typical controls using a subset of 62 subjects from the Centers of Biomedical Research Excellence (COBRE) data repository. We use brain connections estimated by lsXGC as features for classification. After feature extraction, we perform feature selection by Kendall's tau rank correlation coefficient followed by classification using a support vector machine. As a reference method, we compare our results with cross-correlation, typically used in the literature as a standard measure of functional connectivity. We cross-validate 100 different training/test (90%/10%) data split to obtain mean accuracy and a mean Area Under the receiver operating characteristic Curve (AUC) across all tested numbers of features for lsXGC. Our results demonstrate a mean accuracy range of [0.767, 0.940] and a mean AUC range of [0.861, 0.983] for lsXGC. The result of lsXGC is significantly higher than the results obtained with the cross-correlation, namely mean accuracy of [0.721, 0.751] and mean AUC of [0.744, 0.860]. Our results suggest the applicability of lsXGC as a potential biomarker for schizophrenia.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge