Anas Z. Abidin
Large-scale nonlinear Granger causality: A data-driven, multivariate approach to recovering directed networks from short time-series data
Sep 10, 2020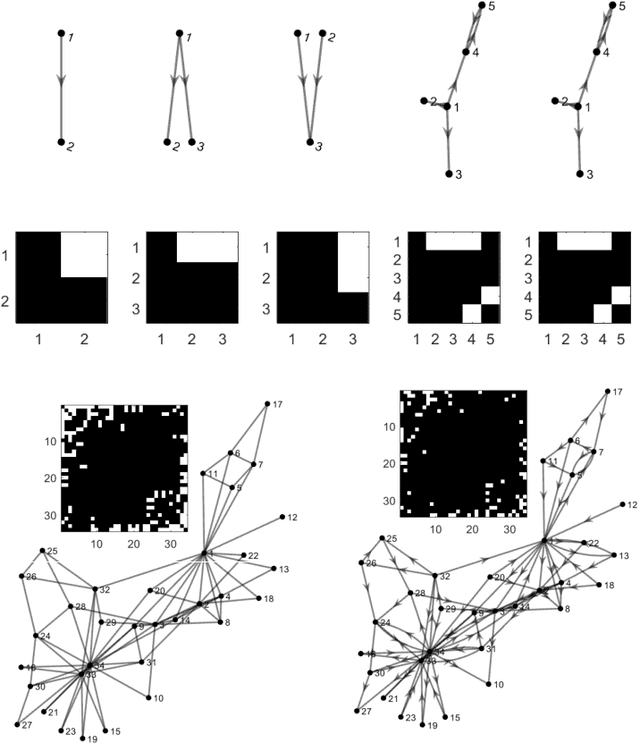
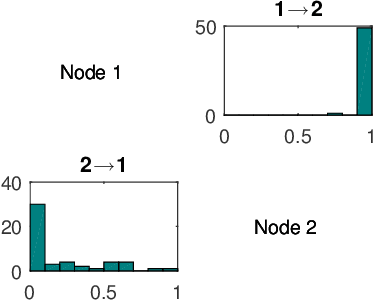
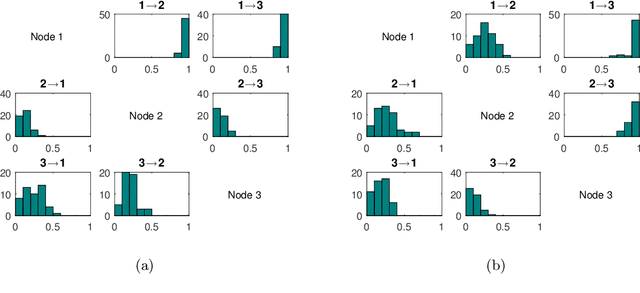
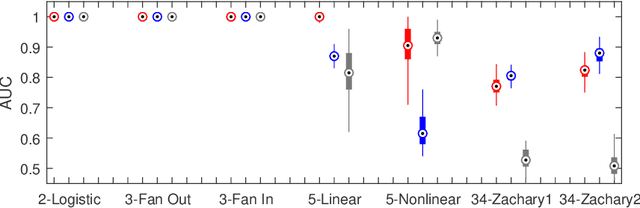
Abstract:To gain insight into complex systems it is a key challenge to infer nonlinear causal directional relations from observational time-series data. Specifically, estimating causal relationships between interacting components in large systems with only short recordings over few temporal observations remains an important, yet unresolved problem. Here, we introduce a large-scale Nonlinear Granger Causality (lsNGC) approach for inferring directional, nonlinear, multivariate causal interactions between system components from short high-dimensional time-series recordings. By modeling interactions with nonlinear state-space transformations from limited observational data, lsNGC identifies casual relations with no explicit a priori assumptions on functional interdependence between component time-series in a computationally efficient manner. Additionally, our method provides a mathematical formulation revealing statistical significance of inferred causal relations. We extensively study the ability of lsNGC to recovering network structure from two-node to thirty-four node chaotic time-series systems. Our results suggest that lsNGC captures meaningful interactions from limited observational data, where it performs favorably when compared to traditionally used methods. Finally, we demonstrate the applicability of lsNGC to estimating causality in large, real-world systems by inferring directional nonlinear, multivariate causal relationships among a large number of relatively short time-series acquired from functional Magnetic Resonance Imaging (fMRI) data of the human brain.
Automated Identification of Thoracic Pathology from Chest Radiographs with Enhanced Training Pipeline
Jun 11, 2020Abstract:Chest x-rays are the most common radiology studies for diagnosing lung and heart disease. Hence, a system for automated pre-reporting of pathologic findings on chest x-rays would greatly enhance radiologists' productivity. To this end, we investigate a deep-learning framework with novel training schemes for classification of different thoracic pathology labels from chest x-rays. We use the currently largest publicly available annotated dataset ChestX-ray14 of 112,120 chest radiographs of 30,805 patients. Each image was annotated with either a 'NoFinding' class, or one or more of 14 thoracic pathology labels. Subjects can have multiple pathologies, resulting in a multi-class, multi-label problem. We encoded labels as binary vectors using k-hot encoding. We study the ResNet34 architecture, pre-trained on ImageNet, where two key modifications were incorporated into the training framework: (1) Stochastic gradient descent with momentum and with restarts using cosine annealing, (2) Variable image sizes for fine-tuning to prevent overfitting. Additionally, we use a heuristic algorithm to select a good learning rate. Learning with restarts was used to avoid local minima. Area Under receiver operating characteristics Curve (AUC) was used to quantitatively evaluate diagnostic quality. Our results are comparable to, or outperform the best results of current state-of-the-art methods with AUCs as follows: Atelectasis:0.81, Cardiomegaly:0.91, Consolidation:0.81, Edema:0.92, Effusion:0.89, Emphysema: 0.92, Fibrosis:0.81, Hernia:0.84, Infiltration:0.73, Mass:0.85, Nodule:0.76, Pleural Thickening:0.81, Pneumonia:0.77, Pneumothorax:0.89 and NoFinding:0.79. Our results suggest that, in addition to using sophisticated network architectures, a good learning rate, scheduler and a robust optimizer can boost performance.
* 6 pages, 1 figure, 2 tables
MRI Tumor Segmentation with Densely Connected 3D CNN
Feb 09, 2018Abstract:Glioma is one of the most common and aggressive types of primary brain tumors. The accurate segmentation of subcortical brain structures is crucial to the study of gliomas in that it helps the monitoring of the progression of gliomas and aids the evaluation of treatment outcomes. However, the large amount of required human labor makes it difficult to obtain the manually segmented Magnetic Resonance Imaging (MRI) data, limiting the use of precise quantitative measurements in the clinical practice. In this work, we try to address this problem by developing a 3D Convolutional Neural Network~(3D CNN) based model to automatically segment gliomas. The major difficulty of our segmentation model comes with the fact that the location, structure, and shape of gliomas vary significantly among different patients. In order to accurately classify each voxel, our model captures multi-scale contextual information by extracting features from two scales of receptive fields. To fully exploit the tumor structure, we propose a novel architecture that hierarchically segments different lesion regions of the necrotic and non-enhancing tumor~(NCR/NET), peritumoral edema~(ED) and GD-enhancing tumor~(ET). Additionally, we utilize densely connected convolutional blocks to further boost the performance. We train our model with a patch-wise training schema to mitigate the class imbalance problem. The proposed method is validated on the BraTS 2017 dataset and it achieves Dice scores of 0.72, 0.83 and 0.81 for the complete tumor, tumor core and enhancing tumor, respectively. These results are comparable to the reported state-of-the-art results, and our method is better than existing 3D-based methods in terms of compactness, time and space efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge