Axel Wismuller
Leveraging Pre-Images to Discover Nonlinear Relationships in Multivariate Environments
Jun 01, 2021

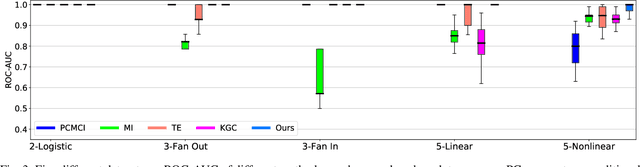

Abstract:Causal discovery, beyond the inference of a network as a collection of connected dots, offers a crucial functionality in scientific discovery using artificial intelligence. The questions that arise in multiple domains, such as physics, physiology, the strategic decision in uncertain environments with multiple agents, climatology, among many others, have roots in causality and reasoning. It became apparent that many real-world temporal observations are nonlinearly related to each other. While the number of observations can be as high as millions of points, the number of temporal samples can be minimal due to ethical or practical reasons, leading to the curse-of-dimensionality in large-scale systems. This paper proposes a novel method using kernel principal component analysis and pre-images to obtain nonlinear dependencies of multivariate time-series data. We show that our method outperforms state-of-the-art causal discovery methods when the observations are restricted by time and are nonlinearly related. Extensive simulations on both real-world and synthetic datasets with various topologies are provided to evaluate our proposed methods.
Large-scale Augmented Granger Causality (lsAGC) for Connectivity Analysis in Complex Systems: From Computer Simulations to Functional MRI (fMRI)
Jan 10, 2021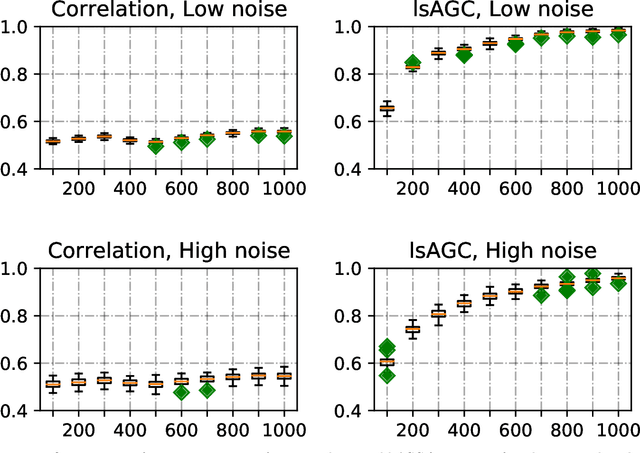
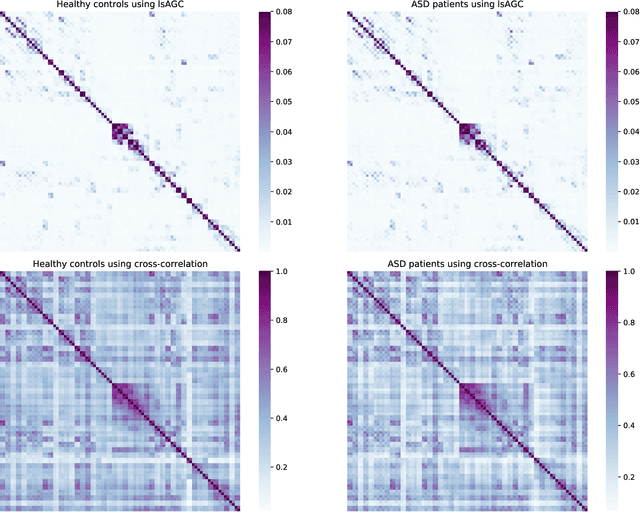
Abstract:We introduce large-scale Augmented Granger Causality (lsAGC) as a method for connectivity analysis in complex systems. The lsAGC algorithm combines dimension reduction with source time-series augmentation and uses predictive time-series modeling for estimating directed causal relationships among time-series. This method is a multivariate approach, since it is capable of identifying the influence of each time-series on any other time-series in the presence of all other time-series of the underlying dynamic system. We quantitatively evaluate the performance of lsAGC on synthetic directional time-series networks with known ground truth. As a reference method, we compare our results with cross-correlation, which is typically used as a standard measure of connectivity in the functional MRI (fMRI) literature. Using extensive simulations for a wide range of time-series lengths and two different signal-to-noise ratios of 5 and 15 dB, lsAGC consistently outperforms cross-correlation at accurately detecting network connections, using Receiver Operator Characteristic Curve (ROC) analysis, across all tested time-series lengths and noise levels. In addition, as an outlook to possible clinical application, we perform a preliminary qualitative analysis of connectivity matrices for fMRI data of Autism Spectrum Disorder (ASD) patients and typical controls, using a subset of 59 subjects of the Autism Brain Imaging Data Exchange II (ABIDE II) data repository. Our results suggest that lsAGC, by extracting sparse connectivity matrices, may be useful for network analysis in complex systems, and may be applicable to clinical fMRI analysis in future research, such as targeting disease-related classification or regression tasks on clinical data.
Large-Scale Extended Granger Causality for Classification of Marijuana Users From Functional MRI
Jan 06, 2021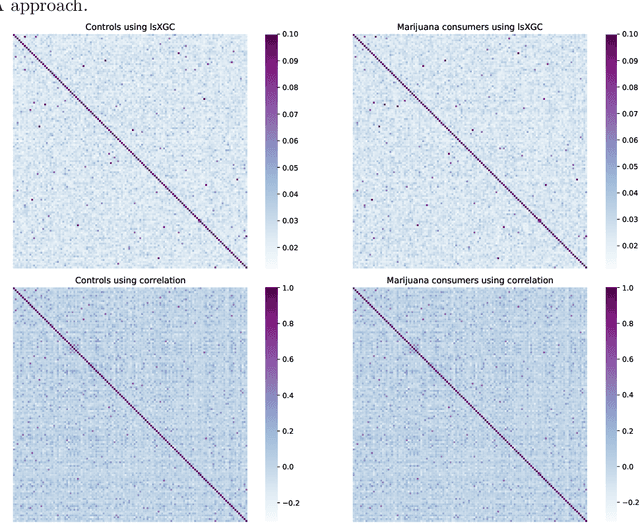
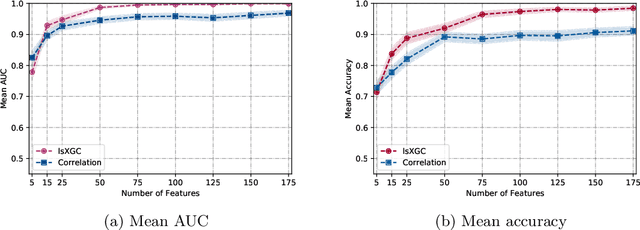
Abstract:It has been shown in the literature that marijuana use is associated with changes in brain network connectivity. We propose large-scale Extended Granger Causality (lsXGC) and investigate whether it can capture such changes using resting-state fMRI. This method combines dimension reduction with source time-series augmentation and uses predictive time-series modeling for estimating directed causal relationships among fMRI time-series. It is a multivariate approach, since it is capable of identifying the interdependence of time-series in the presence of all other time-series of the underlying dynamic system. Here, we investigate whether this model can serve as a biomarker for classifying marijuana users from typical controls using 126 adult subjects with a childhood diagnosis of ADHD from the Addiction Connectome Preprocessed Initiative (ACPI) database. We use brain connections estimated by lsXGC as features for classification. After feature extraction, we perform feature selection by Kendall's-tau rank correlation coefficient followed by classification using a support vector machine. As a reference method, we compare our results with cross-correlation, which is typically used in the literature as a standard measure of functional connectivity. Within a cross-validation scheme of 100 different training/test (90%/10%) data splits, we obtain a mean accuracy range of [0.714, 0.985] and a mean Area Under the receiver operating characteristic Curve (AUC) range of [0.779, 0.999] across all tested numbers of features for lsXGC, which is significantly better than results obtained with cross-correlation, namely mean accuracy of [0.728, 0.912] and mean AUC of [0.825, 0.969]. Our results suggest the applicability of lsXGC as a potential biomarker for marijuana use.
Large-scale kernelized GRANGER causality to infer topology of directed graphs with applications to brain networks
Nov 16, 2020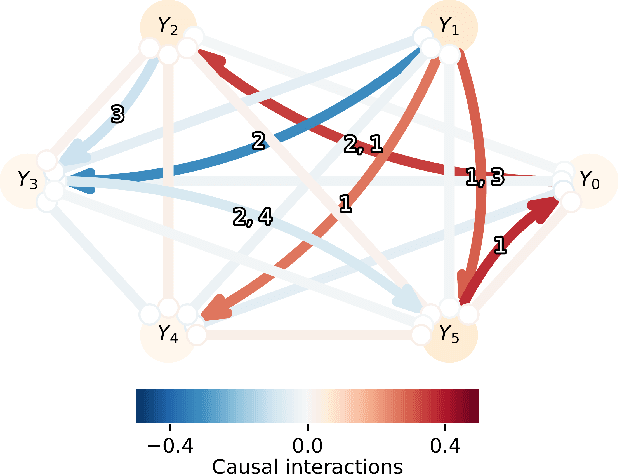
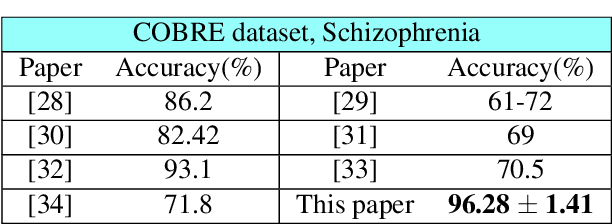
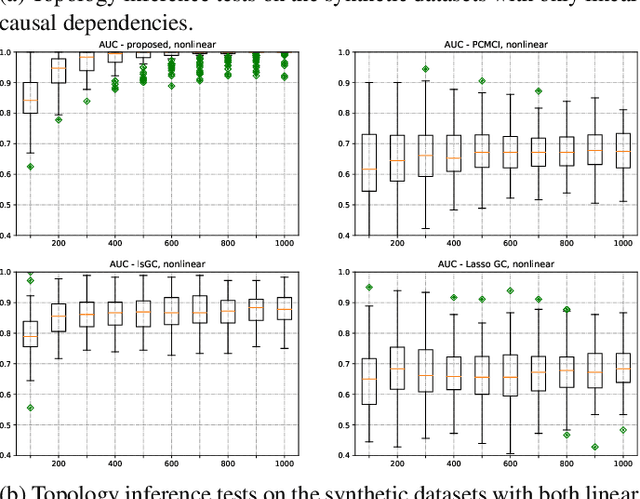
Abstract:Graph topology inference of network processes with co-evolving and interacting time-series is crucial for network studies. Vector autoregressive models (VAR) are popular approaches for topology inference of directed graphs; however, in large networks with short time-series, topology estimation becomes ill-posed. The present paper proposes a novel nonlinearity-preserving topology inference method for directed networks with co-evolving nodal processes that solves the ill-posedness problem. The proposed method, large-scale kernelized Granger causality (lsKGC), uses kernel functions to transform data into a low-dimensional feature space and solves the autoregressive problem in the feature space, then finds the pre-images in the input space to infer the topology. Extensive simulations on synthetic datasets with nonlinear and linear dependencies and known ground-truth demonstrate significant improvement in the Area Under the receiver operating characteristic Curve ( AUC ) of the receiver operating characteristic for network recovery compared to existing methods. Furthermore, tests on real datasets from a functional magnetic resonance imaging (fMRI) study demonstrate 96.3 percent accuracy in diagnosis tasks of schizophrenia patients, which is the highest in the literature with only brain time-series information.
MRI Tumor Segmentation with Densely Connected 3D CNN
Feb 09, 2018Abstract:Glioma is one of the most common and aggressive types of primary brain tumors. The accurate segmentation of subcortical brain structures is crucial to the study of gliomas in that it helps the monitoring of the progression of gliomas and aids the evaluation of treatment outcomes. However, the large amount of required human labor makes it difficult to obtain the manually segmented Magnetic Resonance Imaging (MRI) data, limiting the use of precise quantitative measurements in the clinical practice. In this work, we try to address this problem by developing a 3D Convolutional Neural Network~(3D CNN) based model to automatically segment gliomas. The major difficulty of our segmentation model comes with the fact that the location, structure, and shape of gliomas vary significantly among different patients. In order to accurately classify each voxel, our model captures multi-scale contextual information by extracting features from two scales of receptive fields. To fully exploit the tumor structure, we propose a novel architecture that hierarchically segments different lesion regions of the necrotic and non-enhancing tumor~(NCR/NET), peritumoral edema~(ED) and GD-enhancing tumor~(ET). Additionally, we utilize densely connected convolutional blocks to further boost the performance. We train our model with a patch-wise training schema to mitigate the class imbalance problem. The proposed method is validated on the BraTS 2017 dataset and it achieves Dice scores of 0.72, 0.83 and 0.81 for the complete tumor, tumor core and enhancing tumor, respectively. These results are comparable to the reported state-of-the-art results, and our method is better than existing 3D-based methods in terms of compactness, time and space efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge