Vaibhav Rajan
Segmentation-Aware Latent Diffusion for Satellite Image Super-Resolution: Enabling Smallholder Farm Boundary Delineation
Nov 18, 2025Abstract:Delineating farm boundaries through segmentation of satellite images is a fundamental step in many agricultural applications. The task is particularly challenging for smallholder farms, where accurate delineation requires the use of high resolution (HR) imagery which are available only at low revisit frequencies (e.g., annually). To support more frequent (sub-) seasonal monitoring, HR images could be combined as references (ref) with low resolution (LR) images -- having higher revisit frequency (e.g., weekly) -- using reference-based super-resolution (Ref-SR) methods. However, current Ref-SR methods optimize perceptual quality and smooth over crucial features needed for downstream tasks, and are unable to meet the large scale-factor requirements for this task. Further, previous two-step approaches of SR followed by segmentation do not effectively utilize diverse satellite sources as inputs. We address these problems through a new approach, $\textbf{SEED-SR}$, which uses a combination of conditional latent diffusion models and large-scale multi-spectral, multi-source geo-spatial foundation models. Our key innovation is to bypass the explicit SR task in the pixel space and instead perform SR in a segmentation-aware latent space. This unique approach enables us to generate segmentation maps at an unprecedented 20$\times$ scale factor, and rigorous experiments on two large, real datasets demonstrate up to $\textbf{25.5}$ and $\textbf{12.9}$ relative improvement in instance and semantic segmentation metrics respectively over approaches based on state-of-the-art Ref-SR methods.
WISER: Weak supervISion and supErvised Representation learning to improve drug response prediction in cancer
May 07, 2024Abstract:Cancer, a leading cause of death globally, occurs due to genomic changes and manifests heterogeneously across patients. To advance research on personalized treatment strategies, the effectiveness of various drugs on cells derived from cancers (`cell lines') is experimentally determined in laboratory settings. Nevertheless, variations in the distribution of genomic data and drug responses between cell lines and humans arise due to biological and environmental differences. Moreover, while genomic profiles of many cancer patients are readily available, the scarcity of corresponding drug response data limits the ability to train machine learning models that can predict drug response in patients effectively. Recent cancer drug response prediction methods have largely followed the paradigm of unsupervised domain-invariant representation learning followed by a downstream drug response classification step. Introducing supervision in both stages is challenging due to heterogeneous patient response to drugs and limited drug response data. This paper addresses these challenges through a novel representation learning method in the first phase and weak supervision in the second. Experimental results on real patient data demonstrate the efficacy of our method (WISER) over state-of-the-art alternatives on predicting personalized drug response.
Personalised Drug Identifier for Cancer Treatment with Transformers using Auxiliary Information
Feb 16, 2024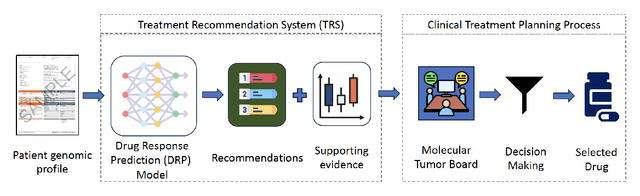
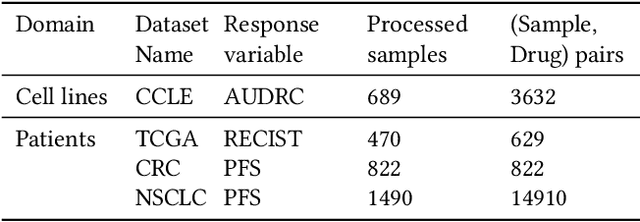
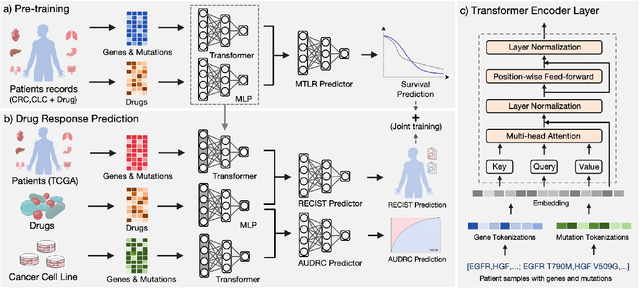
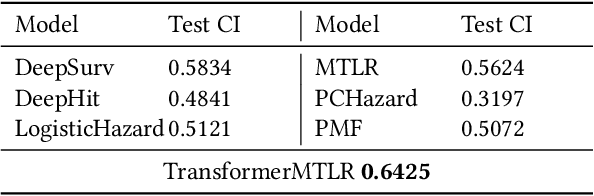
Abstract:Cancer remains a global challenge due to its growing clinical and economic burden. Its uniquely personal manifestation, which makes treatment difficult, has fuelled the quest for personalized treatment strategies. Thus, genomic profiling is increasingly becoming part of clinical diagnostic panels. Effective use of such panels requires accurate drug response prediction (DRP) models, which are challenging to build due to limited labelled patient data. Previous methods to address this problem have used various forms of transfer learning. However, they do not explicitly model the variable length sequential structure of the list of mutations in such diagnostic panels. Further, they do not utilize auxiliary information (like patient survival) for model training. We address these limitations through a novel transformer based method, which surpasses the performance of state-of-the-art DRP models on benchmark data. We also present the design of a treatment recommendation system (TRS), which is currently deployed at the National University Hospital, Singapore and is being evaluated in a clinical trial.
Mixture-Models: a one-stop Python Library for Model-based Clustering using various Mixture Models
Feb 08, 2024Abstract:\texttt{Mixture-Models} is an open-source Python library for fitting Gaussian Mixture Models (GMM) and their variants, such as Parsimonious GMMs, Mixture of Factor Analyzers, MClust models, Mixture of Student's t distributions, etc. It streamlines the implementation and analysis of these models using various first/second order optimization routines such as Gradient Descent and Newton-CG through automatic differentiation (AD) tools. This helps in extending these models to high-dimensional data, which is first of its kind among Python libraries. The library provides user-friendly model evaluation tools, such as BIC, AIC, and log-likelihood estimation. The source-code is licensed under MIT license and can be accessed at \url{https://github.com/kasakh/Mixture-Models}. The package is highly extensible, allowing users to incorporate new distributions and optimization techniques with ease. We conduct a large scale simulation to compare the performance of various gradient based approaches against Expectation Maximization on a wide range of settings and identify the corresponding best suited approach.
A Joint-Reasoning based Disease Q&A System
Jan 06, 2024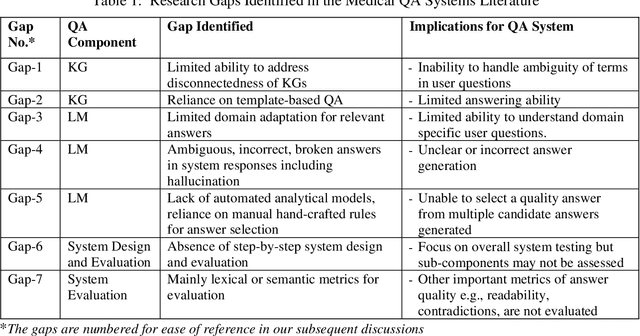
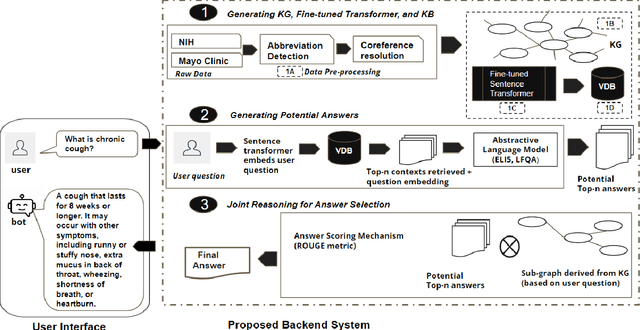
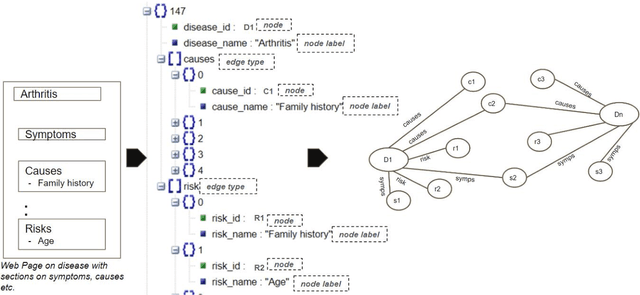
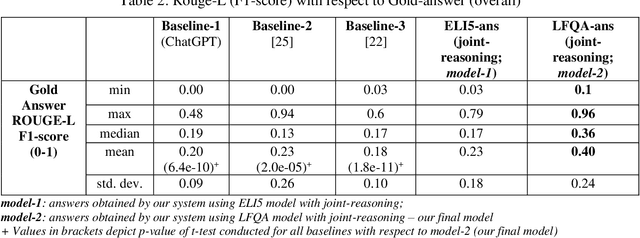
Abstract:Medical question answer (QA) assistants respond to lay users' health-related queries by synthesizing information from multiple sources using natural language processing and related techniques. They can serve as vital tools to alleviate issues of misinformation, information overload, and complexity of medical language, thus addressing lay users' information needs while reducing the burden on healthcare professionals. QA systems, the engines of such assistants, have typically used either language models (LMs) or knowledge graphs (KG), though the approaches could be complementary. LM-based QA systems excel at understanding complex questions and providing well-formed answers, but are prone to factual mistakes. KG-based QA systems, which represent facts well, are mostly limited to answering short-answer questions with pre-created templates. While a few studies have jointly used LM and KG approaches for text-based QA, this was done to answer multiple-choice questions. Extant QA systems also have limitations in terms of automation and performance. We address these challenges by designing a novel, automated disease QA system which effectively utilizes both LM and KG techniques through a joint-reasoning approach to answer disease-related questions appropriate for lay users. Our evaluation of the system using a range of quality metrics demonstrates its efficacy over benchmark systems, including the popular ChatGPT.
ExpertNet: A Symbiosis of Classification and Clustering
Jan 17, 2022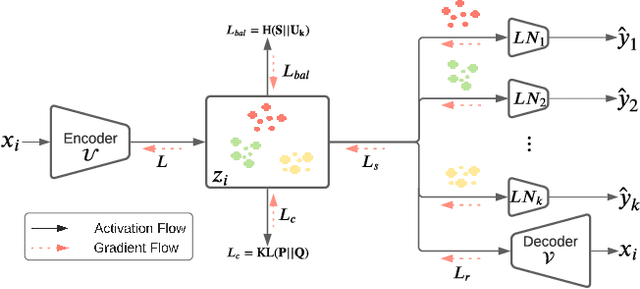
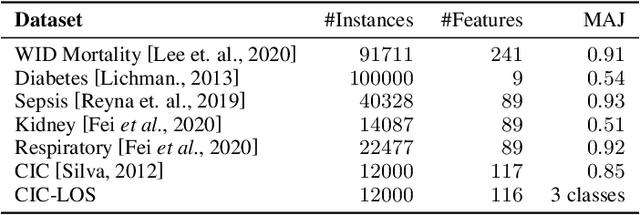

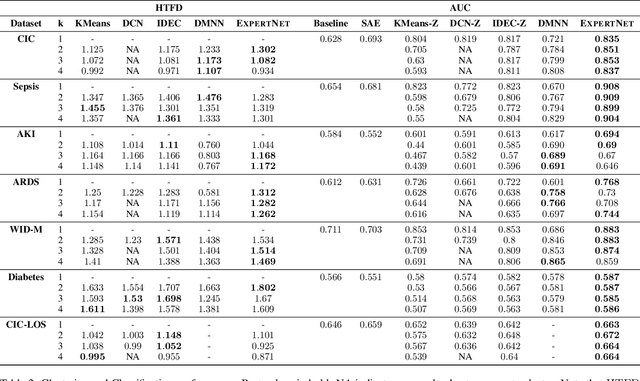
Abstract:A widely used paradigm to improve the generalization performance of high-capacity neural models is through the addition of auxiliary unsupervised tasks during supervised training. Tasks such as similarity matching and input reconstruction have been shown to provide a beneficial regularizing effect by guiding representation learning. Real data often has complex underlying structures and may be composed of heterogeneous subpopulations that are not learned well with current approaches. In this work, we design ExpertNet, which uses novel training strategies to learn clustered latent representations and leverage them by effectively combining cluster-specific classifiers. We theoretically analyze the effect of clustering on its generalization gap, and empirically show that clustered latent representations from ExpertNet lead to disentangling the intrinsic structure and improvement in classification performance. ExpertNet also meets an important real-world need where classifiers need to be tailored for distinct subpopulations, such as in clinical risk models. We demonstrate the superiority of ExpertNet over state-of-the-art methods on 6 large clinical datasets, where our approach leads to valuable insights on group-specific risks.
RepBin: Constraint-based Graph Representation Learning for Metagenomic Binning
Dec 22, 2021



Abstract:Mixed communities of organisms are found in many environments (from the human gut to marine ecosystems) and can have profound impact on human health and the environment. Metagenomics studies the genomic material of such communities through high-throughput sequencing that yields DNA subsequences for subsequent analysis. A fundamental problem in the standard workflow, called binning, is to discover clusters, of genomic subsequences, associated with the unknown constituent organisms. Inherent noise in the subsequences, various biological constraints that need to be imposed on them and the skewed cluster size distribution exacerbate the difficulty of this unsupervised learning problem. In this paper, we present a new formulation using a graph where the nodes are subsequences and edges represent homophily information. In addition, we model biological constraints providing heterophilous signal about nodes that cannot be clustered together. We solve the binning problem by developing new algorithms for (i) graph representation learning that preserves both homophily relations and heterophily constraints (ii) constraint-based graph clustering method that addresses the problems of skewed cluster size distribution. Extensive experiments, on real and synthetic datasets, demonstrate that our approach, called RepBin, outperforms a wide variety of competing methods. Our constraint-based graph representation learning and clustering methods, that may be useful in other domains as well, advance the state-of-the-art in both metagenomics binning and graph representation learning.
Multi-way Clustering and Discordance Analysis through Deep Collective Matrix Tri-Factorization
Sep 27, 2021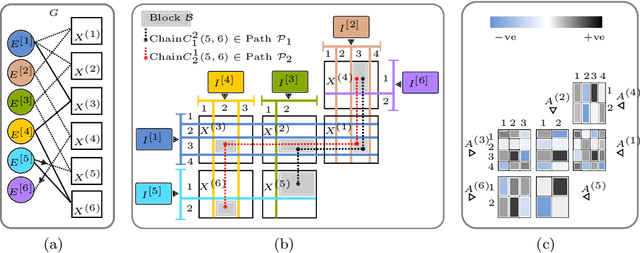
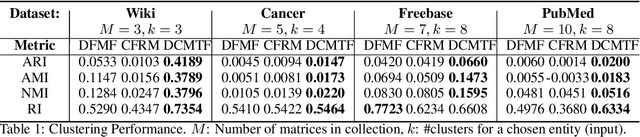
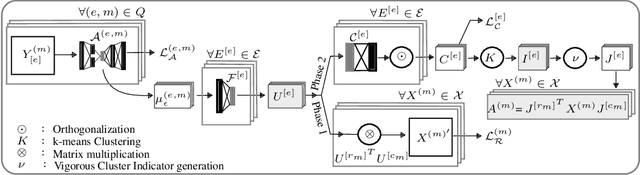
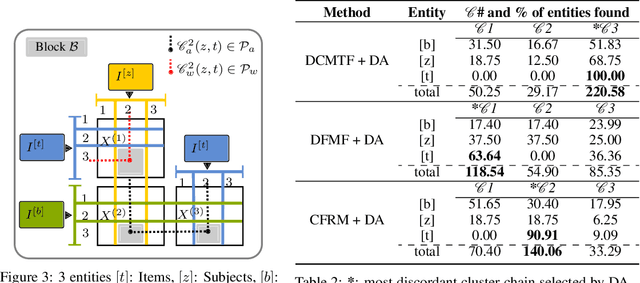
Abstract:Heterogeneous multi-typed, multimodal relational data is increasingly available in many domains and their exploratory analysis poses several challenges. We advance the state-of-the-art in neural unsupervised learning to analyze such data. We design the first neural method for collective matrix tri-factorization of arbitrary collections of matrices to perform spectral clustering of all constituent entities and learn cluster associations. Experiments on benchmark datasets demonstrate its efficacy over previous non-neural approaches. Leveraging signals from multi-way clustering and collective matrix completion we design a unique technique, called Discordance Analysis, to reveal information discrepancies across subsets of matrices in a collection with respect to two entities. We illustrate its utility in quality assessment of knowledge bases and in improving representation learning.
Exact Pareto Optimal Search for Multi-Task Learning: Touring the Pareto Front
Aug 02, 2021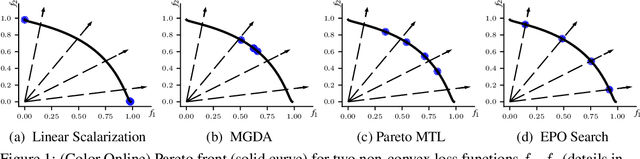
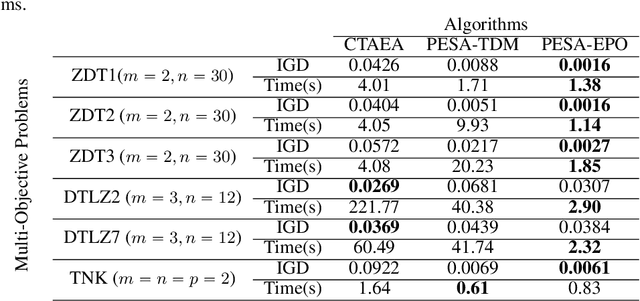
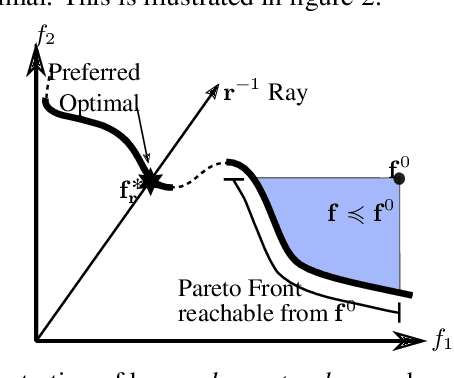
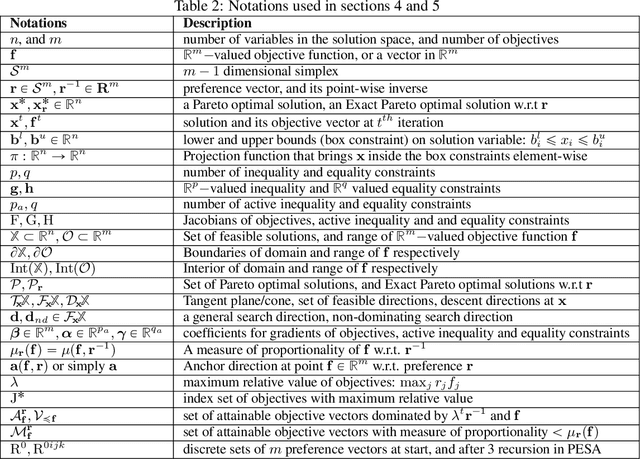
Abstract:Multi-Task Learning (MTL) is a well-established paradigm for training deep neural network models for multiple correlated tasks. Often the task objectives conflict, requiring trade-offs between them during model building. In such cases, MTL models can use gradient-based multi-objective optimization (MOO) to find one or more Pareto optimal solutions. A common requirement in MTL applications is to find an {\it Exact} Pareto optimal (EPO) solution, which satisfies user preferences with respect to task-specific objective functions. Further, to improve model generalization, various constraints on the weights may need to be enforced during training. Addressing these requirements is challenging because it requires a search direction that allows descent not only towards the Pareto front but also towards the input preference, within the constraints imposed and in a manner that scales to high-dimensional gradients. We design and theoretically analyze such search directions and develop the first scalable algorithm, with theoretical guarantees of convergence, to find an EPO solution, including when box and equality constraints are imposed. Our unique method combines multiple gradient descent with carefully controlled ascent to traverse the Pareto front in a principled manner, making it robust to initialization. This also facilitates systematic exploration of the Pareto front, that we utilize to approximate the Pareto front for multi-criteria decision-making. Empirical results show that our algorithm outperforms competing methods on benchmark MTL datasets and MOO problems.
CAC: A Clustering Based Framework for Classification
Feb 23, 2021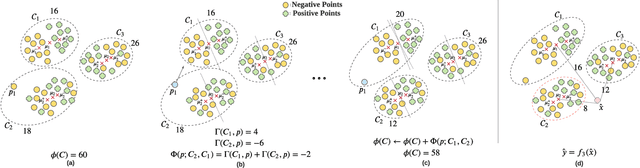
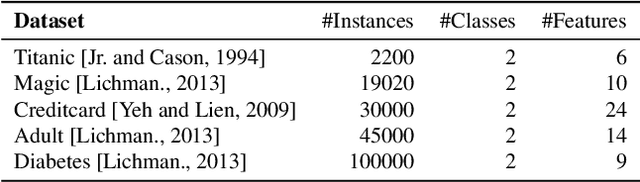
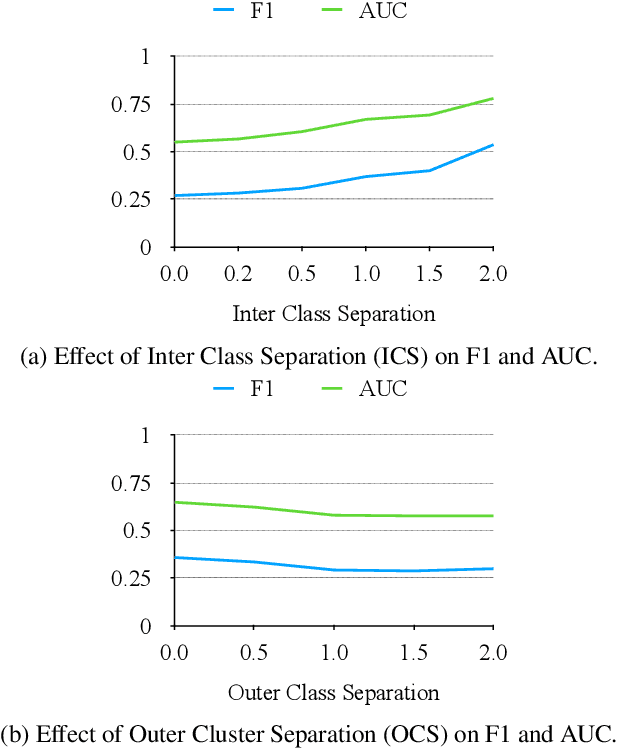
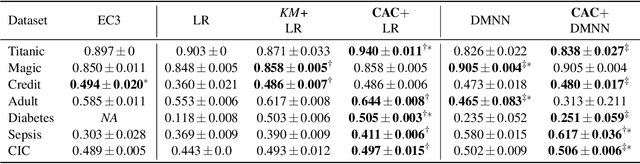
Abstract:In data containing heterogeneous subpopulations, classification performance benefits from incorporating the knowledge of cluster structure in the classifier. Previous methods for such combined clustering and classification either are classifier-specific and not generic or independently perform clustering and classifier training, which may not form clusters that can potentially benefit classifier performance. The question of how to perform clustering to improve the performance of classifiers trained on the clusters has received scant attention in previous literature despite its importance in several real-world applications. In this paper, we theoretically analyze when and how clustering may help in obtaining accurate classifiers. We design a simple, efficient, and generic framework called Classification Aware Clustering (CAC), to find clusters that are well suited for being used as training datasets by classifiers for each underlying subpopulation. Our experiments on synthetic and real benchmark datasets demonstrate the efficacy of CAC over previous methods for combined clustering and classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge