Hansheng Xue
Modeling User Intent Beyond Trigger: Incorporating Uncertainty for Trigger-Induced Recommendation
Aug 07, 2024



Abstract:To cater to users' desire for an immersive browsing experience, numerous e-commerce platforms provide various recommendation scenarios, with a focus on Trigger-Induced Recommendation (TIR) tasks. However, the majority of current TIR methods heavily rely on the trigger item to understand user intent, lacking a higher-level exploration and exploitation of user intent (e.g., popular items and complementary items), which may result in an overly convergent understanding of users' short-term intent and can be detrimental to users' long-term purchasing experiences. Moreover, users' short-term intent shows uncertainty and is affected by various factors such as browsing context and historical behaviors, which poses challenges to user intent modeling. To address these challenges, we propose a novel model called Deep Uncertainty Intent Network (DUIN), comprising three essential modules: i) Explicit Intent Exploit Module extracting explicit user intent using the contrastive learning paradigm; ii) Latent Intent Explore Module exploring latent user intent by leveraging the multi-view relationships between items; iii) Intent Uncertainty Measurement Module offering a distributional estimation and capturing the uncertainty associated with user intent. Experiments on three real-world datasets demonstrate the superior performance of DUIN compared to existing baselines. Notably, DUIN has been deployed across all TIR scenarios in our e-commerce platform, with online A/B testing results conclusively validating its superiority.
Personalised Drug Identifier for Cancer Treatment with Transformers using Auxiliary Information
Feb 16, 2024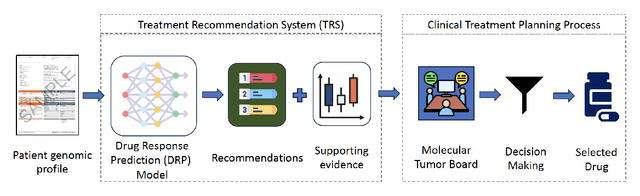
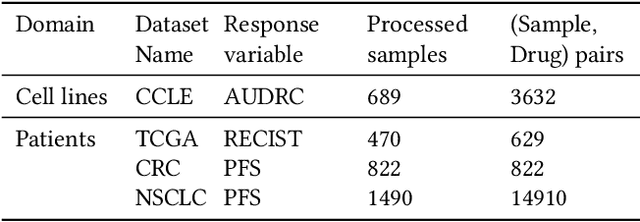
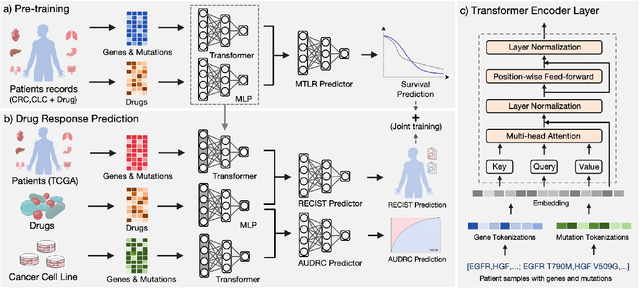
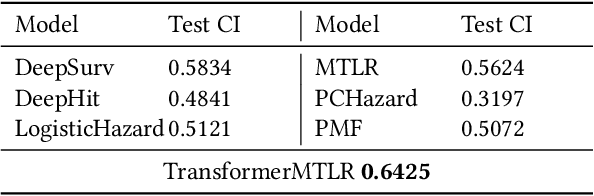
Abstract:Cancer remains a global challenge due to its growing clinical and economic burden. Its uniquely personal manifestation, which makes treatment difficult, has fuelled the quest for personalized treatment strategies. Thus, genomic profiling is increasingly becoming part of clinical diagnostic panels. Effective use of such panels requires accurate drug response prediction (DRP) models, which are challenging to build due to limited labelled patient data. Previous methods to address this problem have used various forms of transfer learning. However, they do not explicitly model the variable length sequential structure of the list of mutations in such diagnostic panels. Further, they do not utilize auxiliary information (like patient survival) for model training. We address these limitations through a novel transformer based method, which surpasses the performance of state-of-the-art DRP models on benchmark data. We also present the design of a treatment recommendation system (TRS), which is currently deployed at the National University Hospital, Singapore and is being evaluated in a clinical trial.
RepBin: Constraint-based Graph Representation Learning for Metagenomic Binning
Dec 22, 2021



Abstract:Mixed communities of organisms are found in many environments (from the human gut to marine ecosystems) and can have profound impact on human health and the environment. Metagenomics studies the genomic material of such communities through high-throughput sequencing that yields DNA subsequences for subsequent analysis. A fundamental problem in the standard workflow, called binning, is to discover clusters, of genomic subsequences, associated with the unknown constituent organisms. Inherent noise in the subsequences, various biological constraints that need to be imposed on them and the skewed cluster size distribution exacerbate the difficulty of this unsupervised learning problem. In this paper, we present a new formulation using a graph where the nodes are subsequences and edges represent homophily information. In addition, we model biological constraints providing heterophilous signal about nodes that cannot be clustered together. We solve the binning problem by developing new algorithms for (i) graph representation learning that preserves both homophily relations and heterophily constraints (ii) constraint-based graph clustering method that addresses the problems of skewed cluster size distribution. Extensive experiments, on real and synthetic datasets, demonstrate that our approach, called RepBin, outperforms a wide variety of competing methods. Our constraint-based graph representation learning and clustering methods, that may be useful in other domains as well, advance the state-of-the-art in both metagenomics binning and graph representation learning.
Multiplex Bipartite Network Embedding using Dual Hypergraph Convolutional Networks
Feb 12, 2021



Abstract:A bipartite network is a graph structure where nodes are from two distinct domains and only inter-domain interactions exist as edges. A large number of network embedding methods exist to learn vectorial node representations from general graphs with both homogeneous and heterogeneous node and edge types, including some that can specifically model the distinct properties of bipartite networks. However, these methods are inadequate to model multiplex bipartite networks (e.g., in e-commerce), that have multiple types of interactions (e.g., click, inquiry, and buy) and node attributes. Most real-world multiplex bipartite networks are also sparse and have imbalanced node distributions that are challenging to model. In this paper, we develop an unsupervised Dual HyperGraph Convolutional Network (DualHGCN) model that scalably transforms the multiplex bipartite network into two sets of homogeneous hypergraphs and uses spectral hypergraph convolutional operators, along with intra- and inter-message passing strategies to promote information exchange within and across domains, to learn effective node embedding. We benchmark DualHGCN using four real-world datasets on link prediction and node classification tasks. Our extensive experiments demonstrate that DualHGCN significantly outperforms state-of-the-art methods, and is robust to varying sparsity levels and imbalanced node distributions.
Modeling Dynamic Heterogeneous Network for Link Prediction using Hierarchical Attention with Temporal RNN
Apr 01, 2020



Abstract:Network embedding aims to learn low-dimensional representations of nodes while capturing structure information of networks. It has achieved great success on many tasks of network analysis such as link prediction and node classification. Most of existing network embedding algorithms focus on how to learn static homogeneous networks effectively. However, networks in the real world are more complex, e.g., networks may consist of several types of nodes and edges (called heterogeneous information) and may vary over time in terms of dynamic nodes and edges (called evolutionary patterns). Limited work has been done for network embedding of dynamic heterogeneous networks as it is challenging to learn both evolutionary and heterogeneous information simultaneously. In this paper, we propose a novel dynamic heterogeneous network embedding method, termed as DyHATR, which uses hierarchical attention to learn heterogeneous information and incorporates recurrent neural networks with temporal attention to capture evolutionary patterns. We benchmark our method on four real-world datasets for the task of link prediction. Experimental results show that DyHATR significantly outperforms several state-of-the-art baselines.
Deep Feature Learning of Multi-Network Topology for Node Classification
Sep 07, 2018



Abstract:Networks are ubiquitous structure that describes complex relationships between different entities in the real world. As a critical component of prediction task over nodes in networks, learning the feature representation of nodes has become one of the most active areas recently. Network Embedding, aiming to learn non-linear and low-dimensional feature representation based on network topology, has been proved to be helpful on tasks of network analysis, especially node classification. For many real-world systems, multiple types of relations are naturally represented by multiple networks. However, existing network embedding methods mainly focus on single network embedding and neglect the information shared among different networks. In this paper, we propose a novel multiple network embedding method based on semisupervised autoencoder, named DeepMNE, which captures complex topological structures of multi-networks and takes the correlation among multi-networks into account. We evaluate DeepMNE on the task of node classification with two real-world datasets. The experimental results demonstrate the superior performance of our method over four state-of-the-art algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge