Thomas Wittenberg
Fraunhofer IIS, Fraunhofer Institute for Integrated Circuits IIS, Friedrich-Alexander-Universität Erlangen-Nürnberg
Instance Segmentation XXL-CT Challenge of a Historic Airplane
Feb 05, 2024Abstract:Instance segmentation of compound objects in XXL-CT imagery poses a unique challenge in non-destructive testing. This complexity arises from the lack of known reference segmentation labels, limited applicable segmentation tools, as well as partially degraded image quality. To asses recent advancements in the field of machine learning-based image segmentation, the "Instance Segmentation XXL-CT Challenge of a Historic Airplane" was conducted. The challenge aimed to explore automatic or interactive instance segmentation methods for an efficient delineation of the different aircraft components, such as screws, rivets, metal sheets or pressure tubes. We report the organization and outcome of this challenge and describe the capabilities and limitations of the submitted segmentation methods.
An annotated instance segmentation XXL-CT dataset from a historic airplane
Dec 16, 2022Abstract:The Me 163 was a Second World War fighter airplane and a result of the German air force secret developments. One of these airplanes is currently owned and displayed in the historic aircraft exhibition of the Deutsches Museum in Munich, Germany. To gain insights with respect to its history, design and state of preservation, a complete CT scan was obtained using an industrial XXL-computer tomography scanner. Using the CT data from the Me 163, all its details can visually be examined at various levels, ranging from the complete hull down to single sprockets and rivets. However, while a trained human observer can identify and interpret the volumetric data with all its parts and connections, a virtual dissection of the airplane and all its different parts would be quite desirable. Nevertheless, this means, that an instance segmentation of all components and objects of interest into disjoint entities from the CT data is necessary. As of currently, no adequate computer-assisted tools for automated or semi-automated segmentation of such XXL-airplane data are available, in a first step, an interactive data annotation and object labeling process has been established. So far, seven 512 x 512 x 512 voxel sub-volumes from the Me 163 airplane have been annotated and labeled, whose results can potentially be used for various new applications in the field of digital heritage, non-destructive testing, or machine-learning. This work describes the data acquisition process of the airplane using an industrial XXL-CT scanner, outlines the interactive segmentation and labeling scheme to annotate sub-volumes of the airplane's CT data, describes and discusses various challenges with respect to interpreting and handling the annotated and labeled data.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022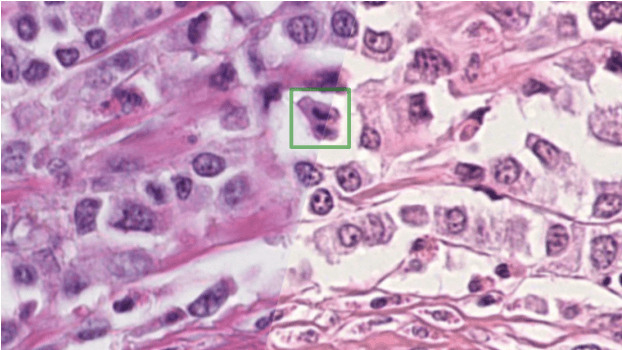
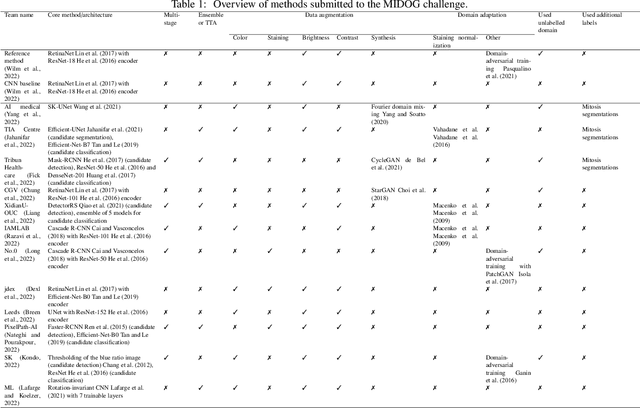
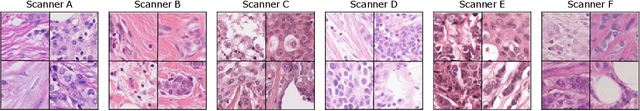
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
MitoDet: Simple and robust mitosis detection
Sep 02, 2021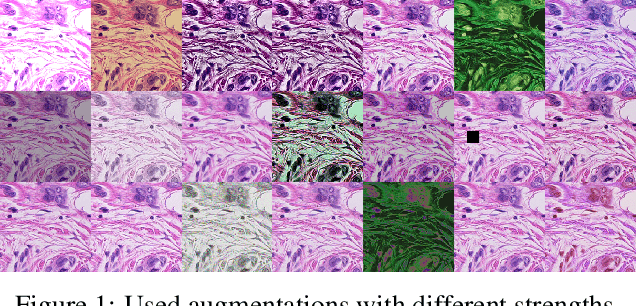
Abstract:Mitotic figure detection is a challenging task in digital pathology that has a direct impact on therapeutic decisions. While automated methods often achieve acceptable results under laboratory conditions, they frequently fail in the clinical deployment phase. This problem can be mainly attributed to a phenomenon called domain shift. An important source of a domain shift is introduced by different microscopes and their camera systems, which noticeably change the color representation of digitized images. In this method description we present our submitted algorithm for the Mitosis Domain Generalization Challenge, which employs a RetinaNet trained with strong data augmentation and achieves an F1 score of 0.7138 on the preliminary test set.
Fast whole-slide cartography in colon cancer histology using superpixels and CNN classification
Jun 30, 2021
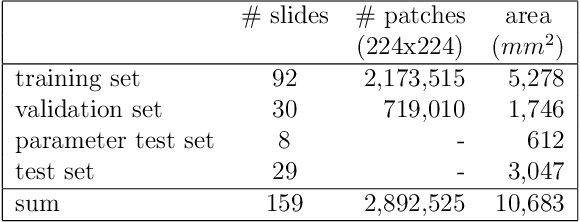
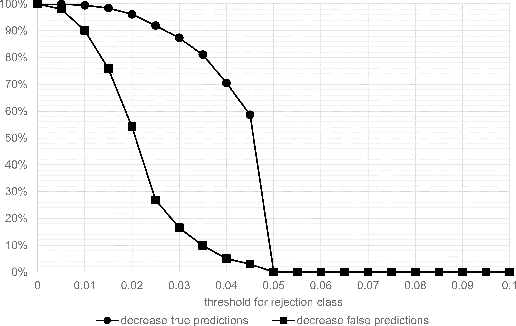
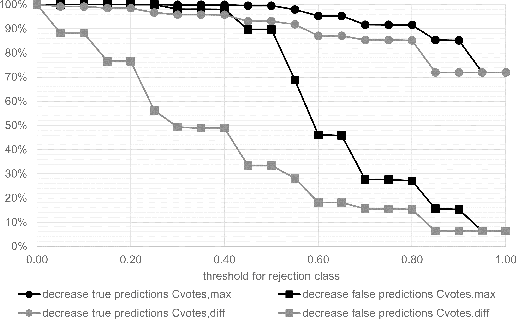
Abstract:Whole-slide-image cartography is the process of automatically detecting and outlining different tissue types in digitized histological specimen. This semantic segmentation provides a basis for many follow-up analyses and can potentially guide subsequent medical decisions. Due to their large size, whole-slide-images typically have to be divided into smaller patches which are then analyzed individually using machine learning-based approaches. Thereby, local dependencies of image regions get lost and since a whole-slide-image comprises many thousands of such patches this process is inherently slow. We propose to subdivide the image into coherent regions prior to classification by grouping visually similar adjacent image pixels into larger segments, i.e. superpixels. Afterwards, only a random subset of patches per superpixel is classified and patch labels are combined into a single superpixel label. The algorithm has been developed and validated on a dataset of 159 hand-annotated whole-slide-images of colon resections and its performance has been compared to a standard patch-based approach. The algorithm shows an average speed-up of 41% on the test data and the overall accuracy is increased from 93.8% to 95.7%. We additionally propose a metric for identifying superpixels with an uncertain classification so they can be excluded from further analysis. Finally, we evaluate two potential medical applications, namely tumor area estimation including tumor invasive margin generation and tumor composition analysis.
Towards a New Science of a Clinical Data Intelligence
Dec 30, 2013
Abstract:In this paper we define Clinical Data Intelligence as the analysis of data generated in the clinical routine with the goal of improving patient care. We define a science of a Clinical Data Intelligence as a data analysis that permits the derivation of scientific, i.e., generalizable and reliable results. We argue that a science of a Clinical Data Intelligence is sensible in the context of a Big Data analysis, i.e., with data from many patients and with complete patient information. We discuss that Clinical Data Intelligence requires the joint efforts of knowledge engineering, information extraction (from textual and other unstructured data), and statistics and statistical machine learning. We describe some of our main results as conjectures and relate them to a recently funded research project involving two major German university hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge