Fast whole-slide cartography in colon cancer histology using superpixels and CNN classification
Paper and Code
Jun 30, 2021
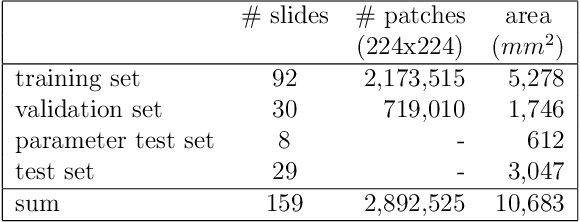
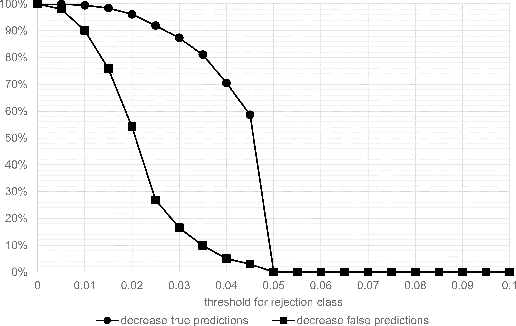
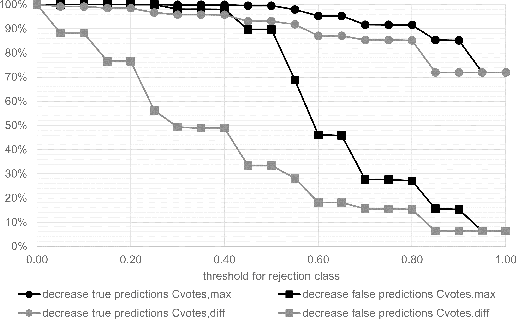
Whole-slide-image cartography is the process of automatically detecting and outlining different tissue types in digitized histological specimen. This semantic segmentation provides a basis for many follow-up analyses and can potentially guide subsequent medical decisions. Due to their large size, whole-slide-images typically have to be divided into smaller patches which are then analyzed individually using machine learning-based approaches. Thereby, local dependencies of image regions get lost and since a whole-slide-image comprises many thousands of such patches this process is inherently slow. We propose to subdivide the image into coherent regions prior to classification by grouping visually similar adjacent image pixels into larger segments, i.e. superpixels. Afterwards, only a random subset of patches per superpixel is classified and patch labels are combined into a single superpixel label. The algorithm has been developed and validated on a dataset of 159 hand-annotated whole-slide-images of colon resections and its performance has been compared to a standard patch-based approach. The algorithm shows an average speed-up of 41% on the test data and the overall accuracy is increased from 93.8% to 95.7%. We additionally propose a metric for identifying superpixels with an uncertain classification so they can be excluded from further analysis. Finally, we evaluate two potential medical applications, namely tumor area estimation including tumor invasive margin generation and tumor composition analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge