Serop Baghdadlian
Explain to Not Forget: Defending Against Catastrophic Forgetting with XAI
May 11, 2022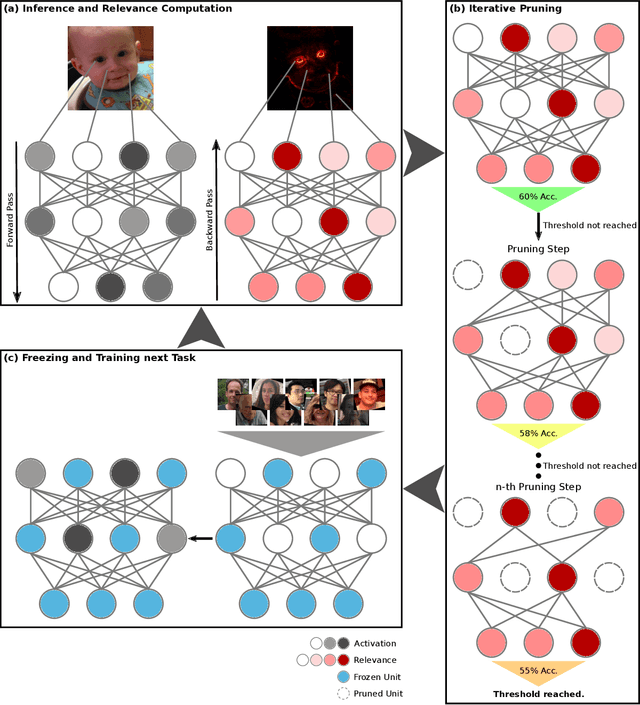
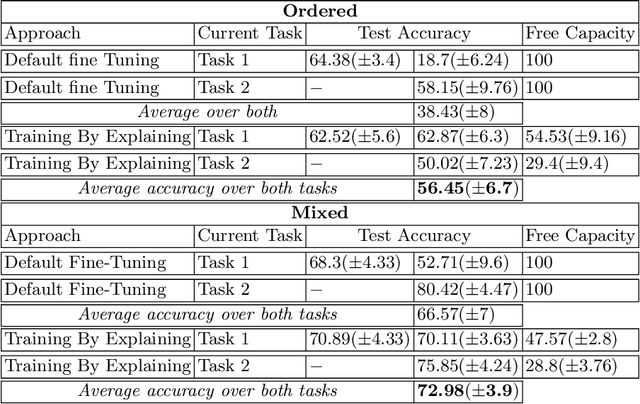
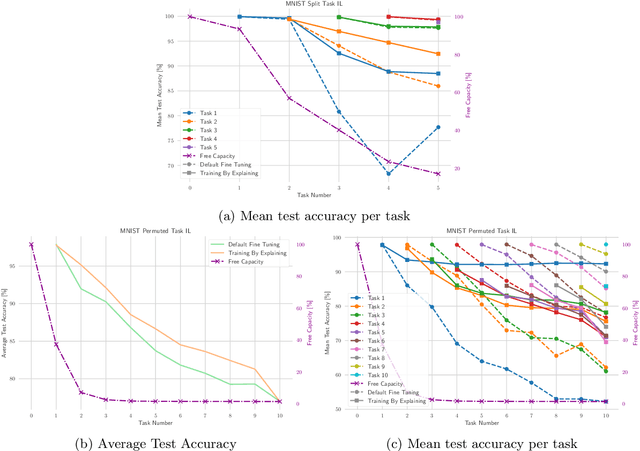
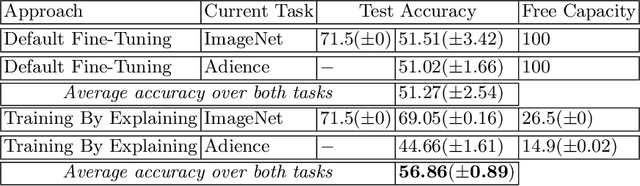
Abstract:The ability to continuously process and retain new information like we do naturally as humans is a feat that is highly sought after when training neural networks. Unfortunately, the traditional optimization algorithms often require large amounts of data available during training time and updates wrt. new data are difficult after the training process has been completed. In fact, when new data or tasks arise, previous progress may be lost as neural networks are prone to catastrophic forgetting. Catastrophic forgetting describes the phenomenon when a neural network completely forgets previous knowledge when given new information. We propose a novel training algorithm called training by explaining in which we leverage Layer-wise Relevance Propagation in order to retain the information a neural network has already learned in previous tasks when training on new data. The method is evaluated on a range of benchmark datasets as well as more complex data. Our method not only successfully retains the knowledge of old tasks within the neural networks but does so more resource-efficiently than other state-of-the-art solutions.
Fast whole-slide cartography in colon cancer histology using superpixels and CNN classification
Jun 30, 2021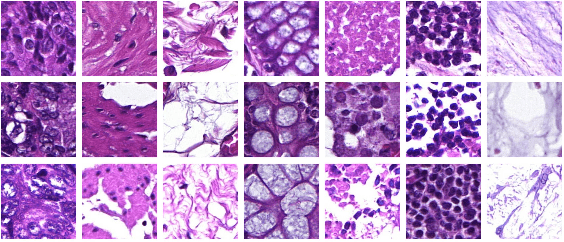
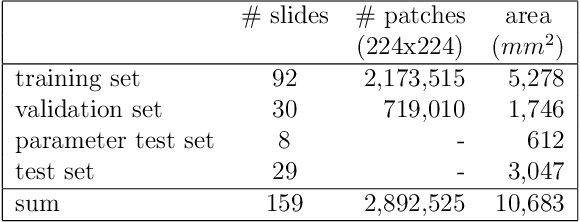
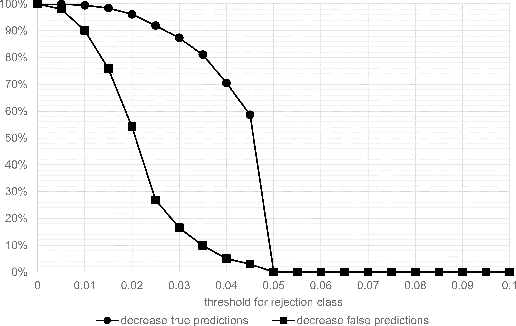
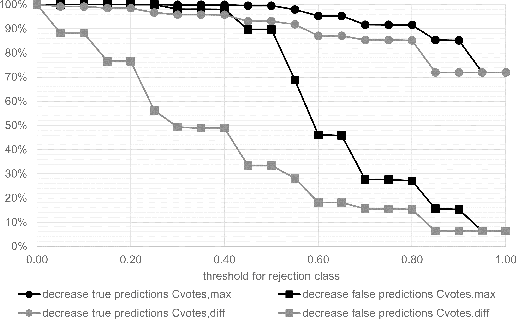
Abstract:Whole-slide-image cartography is the process of automatically detecting and outlining different tissue types in digitized histological specimen. This semantic segmentation provides a basis for many follow-up analyses and can potentially guide subsequent medical decisions. Due to their large size, whole-slide-images typically have to be divided into smaller patches which are then analyzed individually using machine learning-based approaches. Thereby, local dependencies of image regions get lost and since a whole-slide-image comprises many thousands of such patches this process is inherently slow. We propose to subdivide the image into coherent regions prior to classification by grouping visually similar adjacent image pixels into larger segments, i.e. superpixels. Afterwards, only a random subset of patches per superpixel is classified and patch labels are combined into a single superpixel label. The algorithm has been developed and validated on a dataset of 159 hand-annotated whole-slide-images of colon resections and its performance has been compared to a standard patch-based approach. The algorithm shows an average speed-up of 41% on the test data and the overall accuracy is increased from 93.8% to 95.7%. We additionally propose a metric for identifying superpixels with an uncertain classification so they can be excluded from further analysis. Finally, we evaluate two potential medical applications, namely tumor area estimation including tumor invasive margin generation and tumor composition analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge