Jakob Dexl
Post-Training Network Compression for 3D Medical Image Segmentation: Reducing Computational Efforts via Tucker Decomposition
Apr 15, 2024Abstract:We address the computational barrier of deploying advanced deep learning segmentation models in clinical settings by studying the efficacy of network compression through tensor decomposition. We propose a post-training Tucker factorization that enables the decomposition of pre-existing models to reduce computational requirements without impeding segmentation accuracy. We applied Tucker decomposition to the convolutional kernels of the TotalSegmentator (TS) model, an nnU-Net model trained on a comprehensive dataset for automatic segmentation of 117 anatomical structures. Our approach reduced the floating-point operations (FLOPs) and memory required during inference, offering an adjustable trade-off between computational efficiency and segmentation quality. This study utilized the publicly available TS dataset, employing various downsampling factors to explore the relationship between model size, inference speed, and segmentation performance. The application of Tucker decomposition to the TS model substantially reduced the model parameters and FLOPs across various compression rates, with limited loss in segmentation accuracy. We removed up to 88% of the model's parameters with no significant performance changes in the majority of classes after fine-tuning. Practical benefits varied across different graphics processing unit (GPU) architectures, with more distinct speed-ups on less powerful hardware. Post-hoc network compression via Tucker decomposition presents a viable strategy for reducing the computational demand of medical image segmentation models without substantially sacrificing accuracy. This approach enables the broader adoption of advanced deep learning technologies in clinical practice, offering a way to navigate the constraints of hardware capabilities.
ChatGPT Makes Medicine Easy to Swallow: An Exploratory Case Study on Simplified Radiology Reports
Dec 30, 2022Abstract:The release of ChatGPT, a language model capable of generating text that appears human-like and authentic, has gained significant attention beyond the research community. We expect that the convincing performance of ChatGPT incentivizes users to apply it to a variety of downstream tasks, including prompting the model to simplify their own medical reports. To investigate this phenomenon, we conducted an exploratory case study. In a questionnaire, we asked 15 radiologists to assess the quality of radiology reports simplified by ChatGPT. Most radiologists agreed that the simplified reports were factually correct, complete, and not potentially harmful to the patient. Nevertheless, instances of incorrect statements, missed key medical findings, and potentially harmful passages were reported. While further studies are needed, the initial insights of this study indicate a great potential in using large language models like ChatGPT to improve patient-centered care in radiology and other medical domains.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022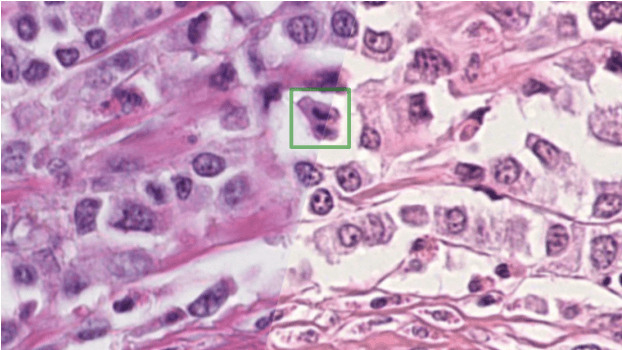
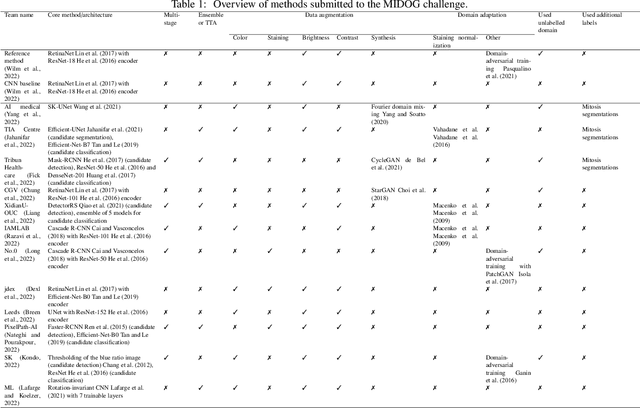
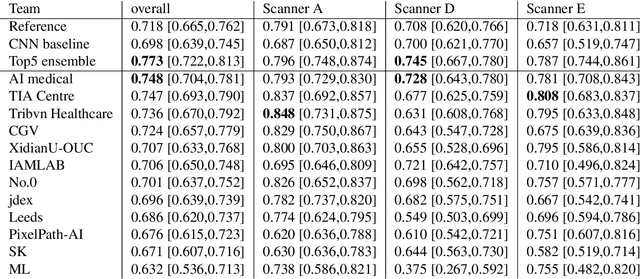
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
MitoDet: Simple and robust mitosis detection
Sep 02, 2021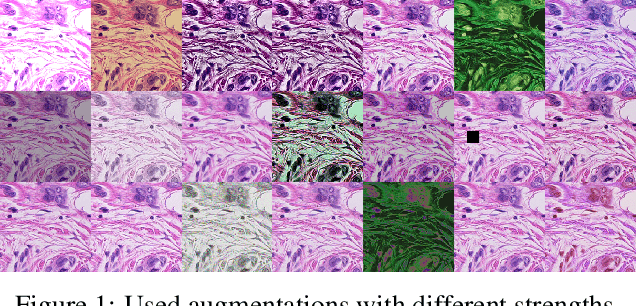
Abstract:Mitotic figure detection is a challenging task in digital pathology that has a direct impact on therapeutic decisions. While automated methods often achieve acceptable results under laboratory conditions, they frequently fail in the clinical deployment phase. This problem can be mainly attributed to a phenomenon called domain shift. An important source of a domain shift is introduced by different microscopes and their camera systems, which noticeably change the color representation of digitized images. In this method description we present our submitted algorithm for the Mitosis Domain Generalization Challenge, which employs a RetinaNet trained with strong data augmentation and achieves an F1 score of 0.7138 on the preliminary test set.
Fast whole-slide cartography in colon cancer histology using superpixels and CNN classification
Jun 30, 2021
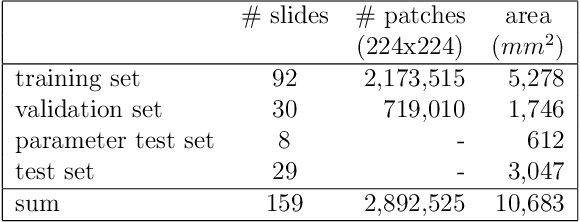
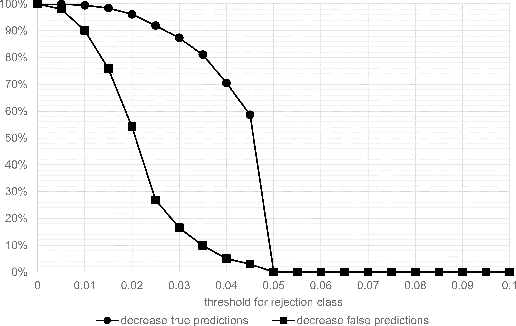
Abstract:Whole-slide-image cartography is the process of automatically detecting and outlining different tissue types in digitized histological specimen. This semantic segmentation provides a basis for many follow-up analyses and can potentially guide subsequent medical decisions. Due to their large size, whole-slide-images typically have to be divided into smaller patches which are then analyzed individually using machine learning-based approaches. Thereby, local dependencies of image regions get lost and since a whole-slide-image comprises many thousands of such patches this process is inherently slow. We propose to subdivide the image into coherent regions prior to classification by grouping visually similar adjacent image pixels into larger segments, i.e. superpixels. Afterwards, only a random subset of patches per superpixel is classified and patch labels are combined into a single superpixel label. The algorithm has been developed and validated on a dataset of 159 hand-annotated whole-slide-images of colon resections and its performance has been compared to a standard patch-based approach. The algorithm shows an average speed-up of 41% on the test data and the overall accuracy is increased from 93.8% to 95.7%. We additionally propose a metric for identifying superpixels with an uncertain classification so they can be excluded from further analysis. Finally, we evaluate two potential medical applications, namely tumor area estimation including tumor invasive margin generation and tumor composition analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge