Tobias Weber
laplax -- Laplace Approximations with JAX
Jul 22, 2025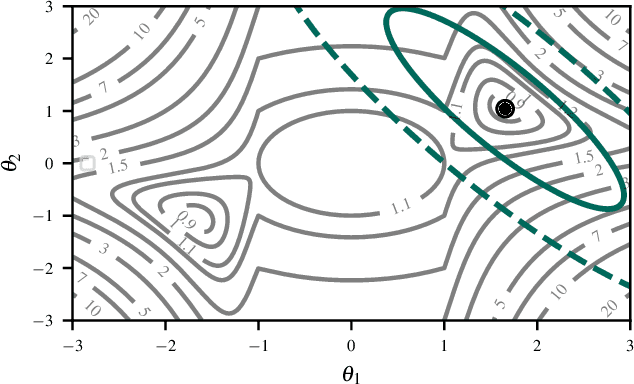
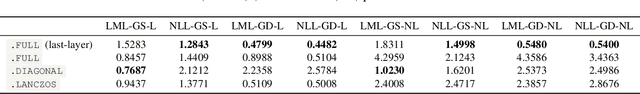
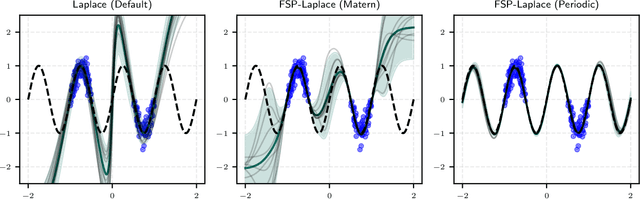

Abstract:The Laplace approximation provides a scalable and efficient means of quantifying weight-space uncertainty in deep neural networks, enabling the application of Bayesian tools such as predictive uncertainty and model selection via Occam's razor. In this work, we introduce laplax, a new open-source Python package for performing Laplace approximations with jax. Designed with a modular and purely functional architecture and minimal external dependencies, laplax offers a flexible and researcher-friendly framework for rapid prototyping and experimentation. Its goal is to facilitate research on Bayesian neural networks, uncertainty quantification for deep learning, and the development of improved Laplace approximation techniques.
Block Graph Neural Networks for tumor heterogeneity prediction
Feb 08, 2025



Abstract:Accurate tumor classification is essential for selecting effective treatments, but current methods have limitations. Standard tumor grading, which categorizes tumors based on cell differentiation, is not recommended as a stand-alone procedure, as some well-differentiated tumors can be malignant. Tumor heterogeneity assessment via single-cell sequencing offers profound insights but can be costly and may still require significant manual intervention. Many existing statistical machine learning methods for tumor data still require complex pre-processing of MRI and histopathological data. In this paper, we propose to build on a mathematical model that simulates tumor evolution (O\.{z}a\'{n}ski (2017)) and generate artificial datasets for tumor classification. Tumor heterogeneity is estimated using normalized entropy, with a threshold to classify tumors as having high or low heterogeneity. Our contributions are threefold: (1) the cut and graph generation processes from the artificial data, (2) the design of tumor features, and (3) the construction of Block Graph Neural Networks (BGNN), a Graph Neural Network-based approach to predict tumor heterogeneity. The experimental results reveal that the combination of the proposed features and models yields excellent results on artificially generated data ($89.67\%$ accuracy on the test data). In particular, in alignment with the emerging trends in AI-assisted grading and spatial transcriptomics, our results suggest that enriching traditional grading methods with birth (e.g., Ki-67 proliferation index) and death markers can improve heterogeneity prediction and enhance tumor classification.
Deep Weight Factorization: Sparse Learning Through the Lens of Artificial Symmetries
Feb 04, 2025



Abstract:Sparse regularization techniques are well-established in machine learning, yet their application in neural networks remains challenging due to the non-differentiability of penalties like the $L_1$ norm, which is incompatible with stochastic gradient descent. A promising alternative is shallow weight factorization, where weights are decomposed into two factors, allowing for smooth optimization of $L_1$-penalized neural networks by adding differentiable $L_2$ regularization to the factors. In this work, we introduce deep weight factorization, extending previous shallow approaches to more than two factors. We theoretically establish equivalence of our deep factorization with non-convex sparse regularization and analyze its impact on training dynamics and optimization. Due to the limitations posed by standard training practices, we propose a tailored initialization scheme and identify important learning rate requirements necessary for training factorized networks. We demonstrate the effectiveness of our deep weight factorization through experiments on various architectures and datasets, consistently outperforming its shallow counterpart and widely used pruning methods.
Linearization Turns Neural Operators into Function-Valued Gaussian Processes
Jun 07, 2024Abstract:Modeling dynamical systems, e.g. in climate and engineering sciences, often necessitates solving partial differential equations. Neural operators are deep neural networks designed to learn nontrivial solution operators of such differential equations from data. As for all statistical models, the predictions of these models are imperfect and exhibit errors. Such errors are particularly difficult to spot in the complex nonlinear behaviour of dynamical systems. We introduce a new framework for approximate Bayesian uncertainty quantification in neural operators using function-valued Gaussian processes. Our approach can be interpreted as a probabilistic analogue of the concept of currying from functional programming and provides a practical yet theoretically sound way to apply the linearized Laplace approximation to neural operators. In a case study on Fourier neural operators, we show that, even for a discretized input, our method yields a Gaussian closure--a structured Gaussian process posterior capturing the uncertainty in the output function of the neural operator, which can be evaluated at an arbitrary set of points. The method adds minimal prediction overhead, can be applied post-hoc without retraining the neural operator, and scales to large models and datasets. We showcase the efficacy of our approach through applications to different types of partial differential equations.
Generalizing Orthogonalization for Models with Non-linearities
May 03, 2024



Abstract:The complexity of black-box algorithms can lead to various challenges, including the introduction of biases. These biases present immediate risks in the algorithms' application. It was, for instance, shown that neural networks can deduce racial information solely from a patient's X-ray scan, a task beyond the capability of medical experts. If this fact is not known to the medical expert, automatic decision-making based on this algorithm could lead to prescribing a treatment (purely) based on racial information. While current methodologies allow for the "orthogonalization" or "normalization" of neural networks with respect to such information, existing approaches are grounded in linear models. Our paper advances the discourse by introducing corrections for non-linearities such as ReLU activations. Our approach also encompasses scalar and tensor-valued predictions, facilitating its integration into neural network architectures. Through extensive experiments, we validate our method's effectiveness in safeguarding sensitive data in generalized linear models, normalizing convolutional neural networks for metadata, and rectifying pre-existing embeddings for undesired attributes.
Post-Training Network Compression for 3D Medical Image Segmentation: Reducing Computational Efforts via Tucker Decomposition
Apr 15, 2024Abstract:We address the computational barrier of deploying advanced deep learning segmentation models in clinical settings by studying the efficacy of network compression through tensor decomposition. We propose a post-training Tucker factorization that enables the decomposition of pre-existing models to reduce computational requirements without impeding segmentation accuracy. We applied Tucker decomposition to the convolutional kernels of the TotalSegmentator (TS) model, an nnU-Net model trained on a comprehensive dataset for automatic segmentation of 117 anatomical structures. Our approach reduced the floating-point operations (FLOPs) and memory required during inference, offering an adjustable trade-off between computational efficiency and segmentation quality. This study utilized the publicly available TS dataset, employing various downsampling factors to explore the relationship between model size, inference speed, and segmentation performance. The application of Tucker decomposition to the TS model substantially reduced the model parameters and FLOPs across various compression rates, with limited loss in segmentation accuracy. We removed up to 88% of the model's parameters with no significant performance changes in the majority of classes after fine-tuning. Practical benefits varied across different graphics processing unit (GPU) architectures, with more distinct speed-ups on less powerful hardware. Post-hoc network compression via Tucker decomposition presents a viable strategy for reducing the computational demand of medical image segmentation models without substantially sacrificing accuracy. This approach enables the broader adoption of advanced deep learning technologies in clinical practice, offering a way to navigate the constraints of hardware capabilities.
Unreading Race: Purging Protected Features from Chest X-ray Embeddings
Nov 02, 2023Abstract:Purpose: To analyze and remove protected feature effects in chest radiograph embeddings of deep learning models. Materials and Methods: An orthogonalization is utilized to remove the influence of protected features (e.g., age, sex, race) in chest radiograph embeddings, ensuring feature-independent results. To validate the efficacy of the approach, we retrospectively study the MIMIC and CheXpert datasets using three pre-trained models, namely a supervised contrastive, a self-supervised contrastive, and a baseline classifier model. Our statistical analysis involves comparing the original versus the orthogonalized embeddings by estimating protected feature influences and evaluating the ability to predict race, age, or sex using the two types of embeddings. Results: Our experiments reveal a significant influence of protected features on predictions of pathologies. Applying orthogonalization removes these feature effects. Apart from removing any influence on pathology classification, while maintaining competitive predictive performance, orthogonalized embeddings further make it infeasible to directly predict protected attributes and mitigate subgroup disparities. Conclusion: The presented work demonstrates the successful application and evaluation of the orthogonalization technique in the domain of chest X-ray classification.
Adversarial Anomaly Detection using Gaussian Priors and Nonlinear Anomaly Scores
Oct 27, 2023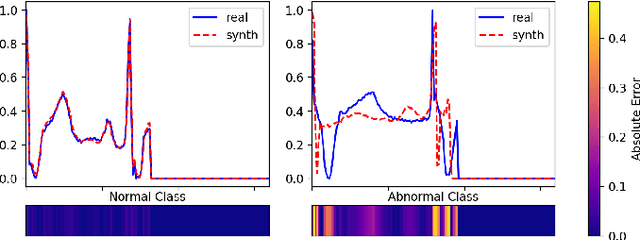
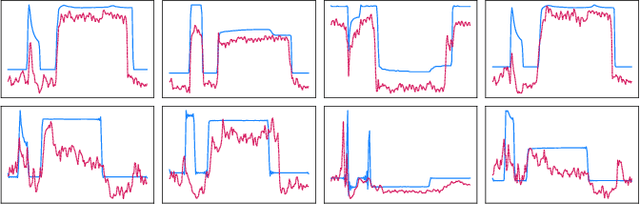
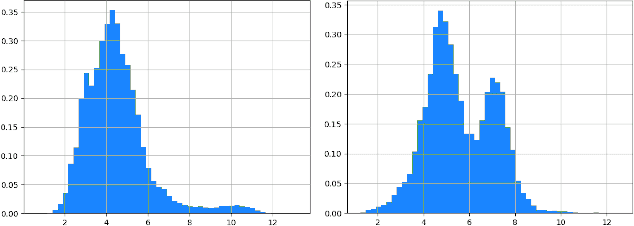
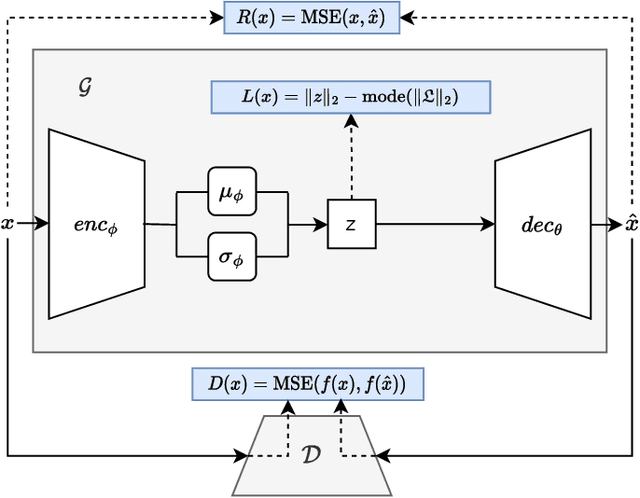
Abstract:Anomaly detection in imbalanced datasets is a frequent and crucial problem, especially in the medical domain where retrieving and labeling irregularities is often expensive. By combining the generative stability of a $\beta$-variational autoencoder (VAE) with the discriminative strengths of generative adversarial networks (GANs), we propose a novel model, $\beta$-VAEGAN. We investigate methods for composing anomaly scores based on the discriminative and reconstructive capabilities of our model. Existing work focuses on linear combinations of these components to determine if data is anomalous. We advance existing work by training a kernelized support vector machine (SVM) on the respective error components to also consider nonlinear relationships. This improves anomaly detection performance, while allowing faster optimization. Lastly, we use the deviations from the Gaussian prior of $\beta$-VAEGAN to form a novel anomaly score component. In comparison to state-of-the-art work, we improve the $F_1$ score during anomaly detection from 0.85 to 0.92 on the widely used MITBIH Arrhythmia Database.
Constrained Probabilistic Mask Learning for Task-specific Undersampled MRI Reconstruction
May 25, 2023



Abstract:Undersampling is a common method in Magnetic Resonance Imaging (MRI) to subsample the number of data points in k-space and thereby reduce acquisition times at the cost of decreased image quality. In this work, we directly learn the undersampling masks to derive task- and domain-specific patterns. To solve this discrete optimization challenge, we propose a general optimization routine called ProM: A fully probabilistic, differentiable, versatile, and model-free framework for mask optimization that enforces acceleration factors through a convex constraint. Analyzing knee, brain, and cardiac MRI datasets with our method, we discover that different anatomic regions reveal distinct optimal undersampling masks. Furthermore, ProM can create undersampling masks that maximize performance in downstream tasks like segmentation with networks trained on fully-sampled MRIs. Even with extreme acceleration factors, ProM yields reasonable performance while being more versatile than existing methods, paving the way for data-driven all-purpose mask generation.
Automated wildlife image classification: An active learning tool for ecological applications
Mar 28, 2023Abstract:Wildlife camera trap images are being used extensively to investigate animal abundance, habitat associations, and behavior, which is complicated by the fact that experts must first classify the images manually. Artificial intelligence systems can take over this task but usually need a large number of already-labeled training images to achieve sufficient performance. This requirement necessitates human expert labor and poses a particular challenge for projects with few cameras or short durations. We propose a label-efficient learning strategy that enables researchers with small or medium-sized image databases to leverage the potential of modern machine learning, thus freeing crucial resources for subsequent analyses. Our methodological proposal is two-fold: (1) We improve current strategies of combining object detection and image classification by tuning the hyperparameters of both models. (2) We provide an active learning (AL) system that allows training deep learning models very efficiently in terms of required human-labeled training images. We supply a software package that enables researchers to use these methods directly and thereby ensure the broad applicability of the proposed framework in ecological practice. We show that our tuning strategy improves predictive performance. We demonstrate how the AL pipeline reduces the amount of pre-labeled data needed to achieve a specific predictive performance and that it is especially valuable for improving out-of-sample predictive performance. We conclude that the combination of tuning and AL increases predictive performance substantially. Furthermore, we argue that our work can broadly impact the community through the ready-to-use software package provided. Finally, the publication of our models tailored to European wildlife data enriches existing model bases mostly trained on data from Africa and North America.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge