Stefan Decker
Unconsciously Forget: Mitigating Memorization; Without Knowing What is being Memorized
Dec 12, 2025Abstract:Recent advances in generative models have demonstrated an exceptional ability to produce highly realistic images. However, previous studies show that generated images often resemble the training data, and this problem becomes more severe as the model size increases. Memorizing training data can lead to legal challenges, including copyright infringement, violations of portrait rights, and trademark violations. Existing approaches to mitigating memorization mainly focus on manipulating the denoising sampling process to steer image embeddings away from the memorized embedding space or employ unlearning methods that require training on datasets containing specific sets of memorized concepts. However, existing methods often incur substantial computational overhead during sampling, or focus narrowly on removing one or more groups of target concepts, imposing a significant limitation on their scalability. To understand and mitigate these problems, our work, UniForget, offers a new perspective on understanding the root cause of memorization. Our work demonstrates that specific parts of the model are responsible for copyrighted content generation. By applying model pruning, we can effectively suppress the probability of generating copyrighted content without targeting specific concepts while preserving the general generative capabilities of the model. Additionally, we show that our approach is both orthogonal and complementary to existing unlearning methods, thereby highlighting its potential to improve current unlearning and de-memorization techniques.
LogicAD: Explainable Anomaly Detection via VLM-based Text Feature Extraction
Jan 08, 2025

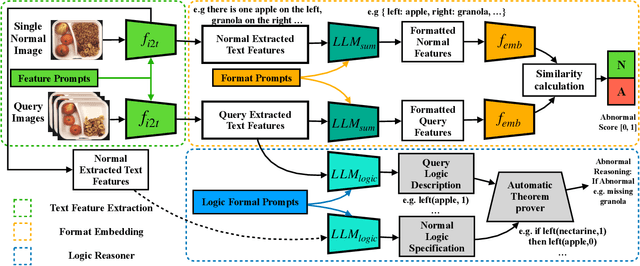
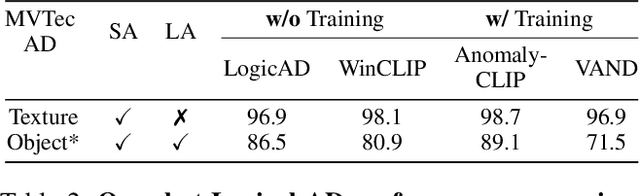
Abstract:Logical image understanding involves interpreting and reasoning about the relationships and consistency within an image's visual content. This capability is essential in applications such as industrial inspection, where logical anomaly detection is critical for maintaining high-quality standards and minimizing costly recalls. Previous research in anomaly detection (AD) has relied on prior knowledge for designing algorithms, which often requires extensive manual annotations, significant computing power, and large amounts of data for training. Autoregressive, multimodal Vision Language Models (AVLMs) offer a promising alternative due to their exceptional performance in visual reasoning across various domains. Despite this, their application to logical AD remains unexplored. In this work, we investigate using AVLMs for logical AD and demonstrate that they are well-suited to the task. Combining AVLMs with format embedding and a logic reasoner, we achieve SOTA performance on public benchmarks, MVTec LOCO AD, with an AUROC of 86.0% and F1-max of 83.7%, along with explanations of anomalies. This significantly outperforms the existing SOTA method by a large margin.
Towards Enabling FAIR Dataspaces Using Large Language Models
Mar 18, 2024Abstract:Dataspaces have recently gained adoption across various sectors, including traditionally less digitized domains such as culture. Leveraging Semantic Web technologies helps to make dataspaces FAIR, but their complexity poses a significant challenge to the adoption of dataspaces and increases their cost. The advent of Large Language Models (LLMs) raises the question of how these models can support the adoption of FAIR dataspaces. In this work, we demonstrate the potential of LLMs in dataspaces with a concrete example. We also derive a research agenda for exploring this emerging field.
From Large Language Models to Knowledge Graphs for Biomarker Discovery in Cancer
Oct 12, 2023Abstract:Domain experts often rely on up-to-date knowledge for apprehending and disseminating specific biological processes that help them design strategies to develop prevention and therapeutic decision-making. A challenging scenario for artificial intelligence (AI) is using biomedical data (e.g., texts, imaging, omics, and clinical) to provide diagnosis and treatment recommendations for cancerous conditions. Data and knowledge about cancer, drugs, genes, proteins, and their mechanism is spread across structured (knowledge bases (KBs)) and unstructured (e.g., scientific articles) sources. A large-scale knowledge graph (KG) can be constructed by integrating these data, followed by extracting facts about semantically interrelated entities and relations. Such KGs not only allow exploration and question answering (QA) but also allow domain experts to deduce new knowledge. However, exploring and querying large-scale KGs is tedious for non-domain users due to a lack of understanding of the underlying data assets and semantic technologies. In this paper, we develop a domain KG to leverage cancer-specific biomarker discovery and interactive QA. For this, a domain ontology called OncoNet Ontology (ONO) is developed to enable semantic reasoning for validating gene-disease relations. The KG is then enriched by harmonizing the ONO, controlled vocabularies, and additional biomedical concepts from scientific articles by employing BioBERT- and SciBERT-based information extraction (IE) methods. Further, since the biomedical domain is evolving, where new findings often replace old ones, without employing up-to-date findings, there is a high chance an AI system exhibits concept drift while providing diagnosis and treatment. Therefore, we finetuned the KG using large language models (LLMs) based on more recent articles and KBs that might not have been seen by the named entity recognition models.
A Biomedical Knowledge Graph for Biomarker Discovery in Cancer
Feb 23, 2023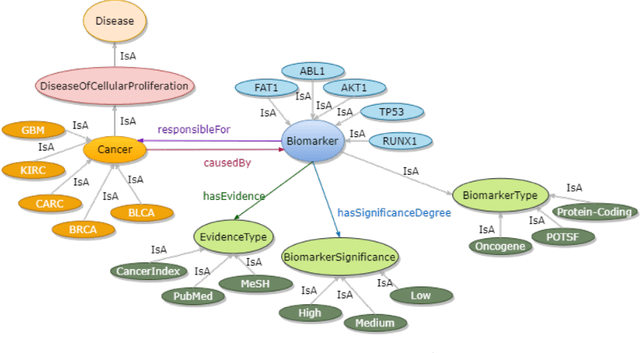
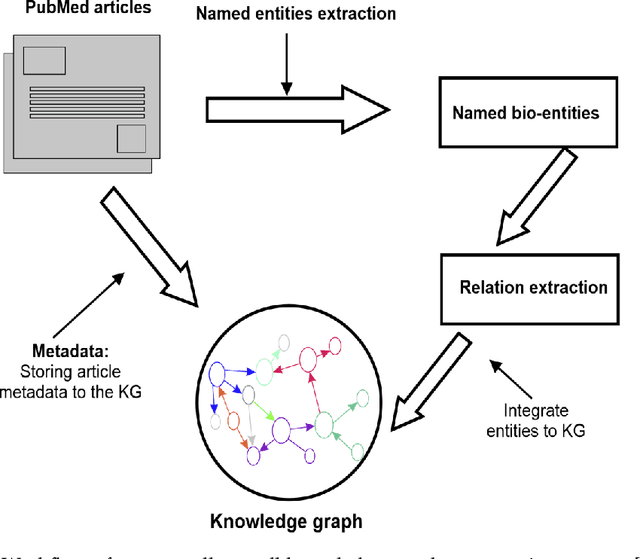

Abstract:Structured and unstructured data and facts about drugs, genes, protein, viruses, and their mechanism are spread across a huge number of scientific articles. These articles are a large-scale knowledge source and can have a huge impact on disseminating knowledge about the mechanisms of certain biological processes. A domain-specific knowledge graph~(KG) is an explicit conceptualization of a specific subject-matter domain represented w.r.t semantically interrelated entities and relations. A KG can be constructed by integrating such facts and data and be used for data integration, exploration, and federated queries. However, exploration and querying large-scale KGs is tedious for certain groups of users due to a lack of knowledge about underlying data assets or semantic technologies. Such a KG will not only allow deducing new knowledge and question answering(QA) but also allows domain experts to explore. Since cross-disciplinary explanations are important for accurate diagnosis, it is important to query the KG to provide interactive explanations about learned biomarkers. Inspired by these, we construct a domain-specific KG, particularly for cancer-specific biomarker discovery. The KG is constructed by integrating cancer-related knowledge and facts from multiple sources. First, we construct a domain-specific ontology, which we call OncoNet Ontology (ONO). The ONO ontology is developed to enable semantic reasoning for verification of the predictions for relations between diseases and genes. The KG is then developed and enriched by harmonizing the ONO, additional metadata schemas, ontologies, controlled vocabularies, and additional concepts from external sources using a BERT-based information extraction method. BioBERT and SciBERT are finetuned with the selected articles crawled from PubMed. We listed down some queries and some examples of QA and deducing knowledge based on the KG.
Explainable AI for Bioinformatics: Methods, Tools, and Applications
Dec 25, 2022Abstract:Artificial intelligence(AI) systems based on deep neural networks (DNNs) and machine learning (ML) algorithms are increasingly used to solve critical problems in bioinformatics, biomedical informatics, and precision medicine. However, complex DNN or ML models that are unavoidably opaque and perceived as black-box methods, may not be able to explain why and how they make certain decisions. Such black-box models are difficult to comprehend not only for targeted users and decision-makers but also for AI developers. Besides, in sensitive areas like healthcare, explainability and accountability are not only desirable properties of AI but also legal requirements -- especially when AI may have significant impacts on human lives. Explainable artificial intelligence (XAI) is an emerging field that aims to mitigate the opaqueness of black-box models and make it possible to interpret how AI systems make their decisions with transparency. An interpretable ML model can explain how it makes predictions and which factors affect the model's outcomes. The majority of state-of-the-art interpretable ML methods have been developed in a domain-agnostic way and originate from computer vision, automated reasoning, or even statistics. Many of these methods cannot be directly applied to bioinformatics problems, without prior customization, extension, and domain adoption. In this paper, we discuss the importance of explainability with a focus on bioinformatics. We analyse and comprehensively overview of model-specific and model-agnostic interpretable ML methods and tools. Via several case studies covering bioimaging, cancer genomics, and biomedical text mining, we show how bioinformatics research could benefit from XAI methods and how they could help improve decision fairness.
Question Answering Over Biological Knowledge Graph via Amazon Alexa
Oct 12, 2022
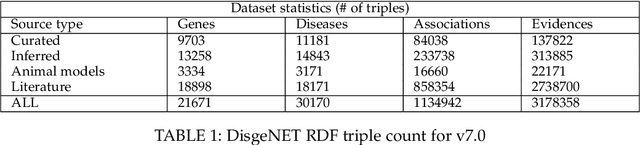
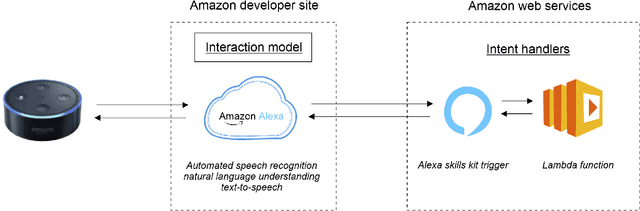
Abstract:Structured and unstructured data and facts about drugs, genes, protein, viruses, and their mechanism are spread across a huge number of scientific articles. These articles are a large-scale knowledge source and can have a huge impact on disseminating knowledge about the mechanisms of certain biological processes. A knowledge graph (KG) can be constructed by integrating such facts and data and be used for data integration, exploration, and federated queries. However, exploration and querying large-scale KGs is tedious for certain groups of users due to a lack of knowledge about underlying data assets or semantic technologies. A question-answering (QA) system allows the answer of natural language questions over KGs automatically using triples contained in a KG. Recently, the use and adaption of digital assistants are getting wider owing to their capability at enabling users to voice commands to control smart systems or devices. This paper is about using Amazon Alexa's voice-enabled interface for QA over KGs. As a proof-of-concept, we use the well-known DisgeNET KG, which contains knowledge covering 1.13 million gene-disease associations between 21,671 genes and 30,170 diseases, disorders, and clinical or abnormal human phenotypes. Our study shows how Alex could be of help to find facts about certain biological entities from large-scale knowledge bases.
Interpreting Black-box Machine Learning Models for High Dimensional Datasets
Aug 29, 2022
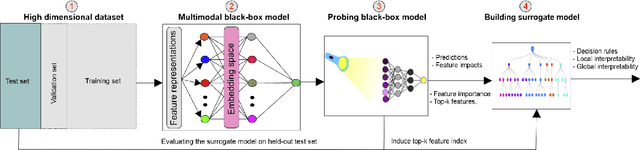


Abstract:Deep neural networks (DNNs) have been shown to outperform traditional machine learning algorithms in a broad variety of application domains due to their effectiveness in modeling intricate problems and handling high-dimensional datasets. Many real-life datasets, however, are of increasingly high dimensionality, where a large number of features may be irrelevant to the task at hand. The inclusion of such features would not only introduce unwanted noise but also increase computational complexity. Furthermore, due to high non-linearity and dependency among a large number of features, DNN models tend to be unavoidably opaque and perceived as black-box methods because of their not well-understood internal functioning. A well-interpretable model can identify statistically significant features and explain the way they affect the model's outcome. In this paper, we propose an efficient method to improve the interpretability of black-box models for classification tasks in the case of high-dimensional datasets. To this end, we first train a black-box model on a high-dimensional dataset to learn the embeddings on which the classification is performed. To decompose the inner working principles of the black-box model and to identify top-k important features, we employ different probing and perturbing techniques. We then approximate the behavior of the black-box model by means of an interpretable surrogate model on the top-k feature space. Finally, we derive decision rules and local explanations from the surrogate model to explain individual decisions. Our approach outperforms and competes with state-of-the-art methods such as TabNet, XGboost, and SHAP-based interpretability techniques when tested on different datasets with varying dimensionality between 50 and 20,000.
Towards General Deep Leakage in Federated Learning
Oct 18, 2021

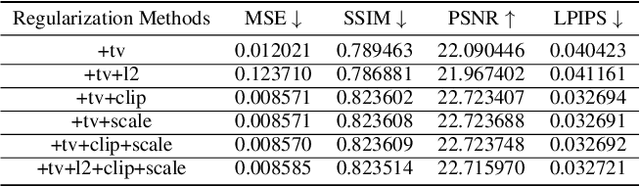
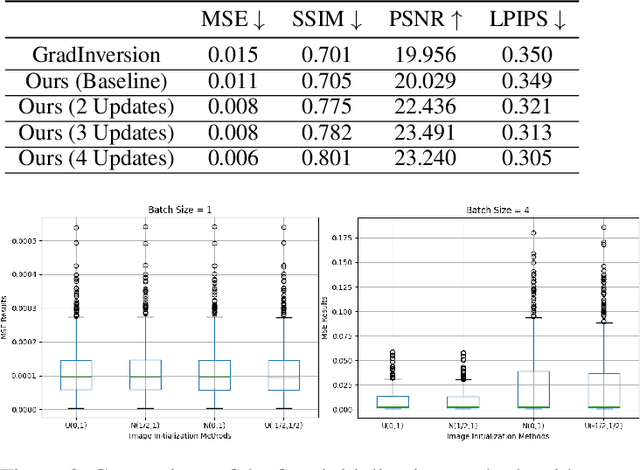
Abstract:Unlike traditional central training, federated learning (FL) improves the performance of the global model by sharing and aggregating local models rather than local data to protect the users' privacy. Although this training approach appears secure, some research has demonstrated that an attacker can still recover private data based on the shared gradient information. This on-the-fly reconstruction attack deserves to be studied in depth because it can occur at any stage of training, whether at the beginning or at the end of model training; no relevant dataset is required and no additional models need to be trained. We break through some unrealistic assumptions and limitations to apply this reconstruction attack in a broader range of scenarios. We propose methods that can reconstruct the training data from shared gradients or weights, corresponding to the FedSGD and FedAvg usage scenarios, respectively. We propose a zero-shot approach to restore labels even if there are duplicate labels in the batch. We study the relationship between the label and image restoration. We find that image restoration fails even if there is only one incorrectly inferred label in the batch; we also find that when batch images have the same label, the corresponding image is restored as a fusion of that class of images. Our approaches are evaluated on classic image benchmarks, including CIFAR-10 and ImageNet. The batch size, image quality, and the adaptability of the label distribution of our approach exceed those of GradInversion, the state-of-the-art.
Secure Evaluation of Knowledge Graph Merging Gain
Feb 26, 2021
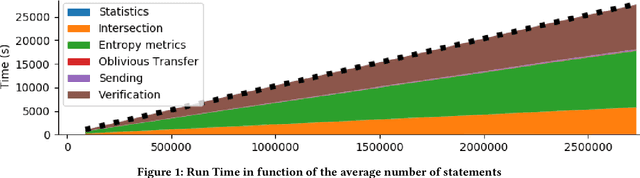


Abstract:Finding out the differences and commonalities between the knowledge of two parties is an important task. Such a comparison becomes necessary, when one party wants to determine how much it is worth to acquire the knowledge of the second party, or similarly when two parties try to determine, whether a collaboration could be beneficial. When these two parties cannot trust each other (for example, due to them being competitors) performing such a comparison is challenging as neither of them would be willing to share any of their assets. This paper addresses this problem for knowledge graphs, without a need for non-disclosure agreements nor a third party during the protocol. During the protocol, the intersection between the two knowledge graphs is determined in a privacy preserving fashion. This is followed by the computation of various metrics, which give an indication of the potential gain from obtaining the other parties knowledge graph, while still keeping the actual knowledge graph contents secret. The protocol makes use of blind signatures and (counting) Bloom filters to reduce the amount of leaked information. Finally, the party who wants to obtain the other's knowledge graph can get a part of such in a way that neither party is able to know beforehand which parts of the graph are obtained (i.e., they cannot choose to only get or share the good parts). After inspection of the quality of this part, the Buyer can decide to proceed with the transaction. The analysis of the protocol indicates that the developed protocol is secure against malicious participants. Further experimental analysis shows that the resource consumption scales linear with the number of statements in the knowledge graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge