Christoph Lange
Towards Enabling FAIR Dataspaces Using Large Language Models
Mar 18, 2024Abstract:Dataspaces have recently gained adoption across various sectors, including traditionally less digitized domains such as culture. Leveraging Semantic Web technologies helps to make dataspaces FAIR, but their complexity poses a significant challenge to the adoption of dataspaces and increases their cost. The advent of Large Language Models (LLMs) raises the question of how these models can support the adoption of FAIR dataspaces. In this work, we demonstrate the potential of LLMs in dataspaces with a concrete example. We also derive a research agenda for exploring this emerging field.
Data Augmentation Scheme for Raman Spectra with Highly Correlated Annotations
Feb 01, 2024Abstract:In biotechnology Raman Spectroscopy is rapidly gaining popularity as a process analytical technology (PAT) that measures cell densities, substrate- and product concentrations. As it records vibrational modes of molecules it provides that information non-invasively in a single spectrum. Typically, partial least squares (PLS) is the model of choice to infer information about variables of interest from the spectra. However, biological processes are known for their complexity where convolutional neural networks (CNN) present a powerful alternative. They can handle non-Gaussian noise and account for beam misalignment, pixel malfunctions or the presence of additional substances. However, they require a lot of data during model training, and they pick up non-linear dependencies in the process variables. In this work, we exploit the additive nature of spectra in order to generate additional data points from a given dataset that have statistically independent labels so that a network trained on such data exhibits low correlations between the model predictions. We show that training a CNN on these generated data points improves the performance on datasets where the annotations do not bear the same correlation as the dataset that was used for model training. This data augmentation technique enables us to reuse spectra as training data for new contexts that exhibit different correlations. The additional data allows for building a better and more robust model. This is of interest in scenarios where large amounts of historical data are available but are currently not used for model training. We demonstrate the capabilities of the proposed method using synthetic spectra of Ralstonia eutropha batch cultivations to monitor substrate, biomass and polyhydroxyalkanoate (PHA) biopolymer concentrations during of the experiments.
Enhancing Data Space Semantic Interoperability through Machine Learning: a Visionary Perspective
Mar 15, 2023Abstract:Our vision paper outlines a plan to improve the future of semantic interoperability in data spaces through the application of machine learning. The use of data spaces, where data is exchanged among members in a self-regulated environment, is becoming increasingly popular. However, the current manual practices of managing metadata and vocabularies in these spaces are time-consuming, prone to errors, and may not meet the needs of all stakeholders. By leveraging the power of machine learning, we believe that semantic interoperability in data spaces can be significantly improved. This involves automatically generating and updating metadata, which results in a more flexible vocabulary that can accommodate the diverse terminologies used by different sub-communities. Our vision for the future of data spaces addresses the limitations of conventional data exchange and makes data more accessible and valuable for all members of the community.
Explainable AI for Bioinformatics: Methods, Tools, and Applications
Dec 25, 2022Abstract:Artificial intelligence(AI) systems based on deep neural networks (DNNs) and machine learning (ML) algorithms are increasingly used to solve critical problems in bioinformatics, biomedical informatics, and precision medicine. However, complex DNN or ML models that are unavoidably opaque and perceived as black-box methods, may not be able to explain why and how they make certain decisions. Such black-box models are difficult to comprehend not only for targeted users and decision-makers but also for AI developers. Besides, in sensitive areas like healthcare, explainability and accountability are not only desirable properties of AI but also legal requirements -- especially when AI may have significant impacts on human lives. Explainable artificial intelligence (XAI) is an emerging field that aims to mitigate the opaqueness of black-box models and make it possible to interpret how AI systems make their decisions with transparency. An interpretable ML model can explain how it makes predictions and which factors affect the model's outcomes. The majority of state-of-the-art interpretable ML methods have been developed in a domain-agnostic way and originate from computer vision, automated reasoning, or even statistics. Many of these methods cannot be directly applied to bioinformatics problems, without prior customization, extension, and domain adoption. In this paper, we discuss the importance of explainability with a focus on bioinformatics. We analyse and comprehensively overview of model-specific and model-agnostic interpretable ML methods and tools. Via several case studies covering bioimaging, cancer genomics, and biomedical text mining, we show how bioinformatics research could benefit from XAI methods and how they could help improve decision fairness.
OncoNetExplainer: Explainable Predictions of Cancer Types Based on Gene Expression Data
Sep 09, 2019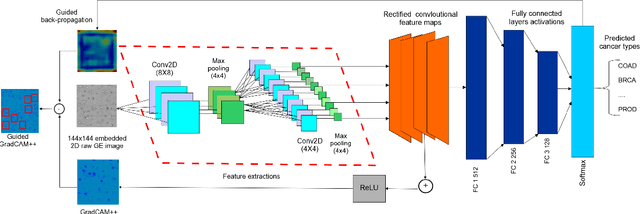
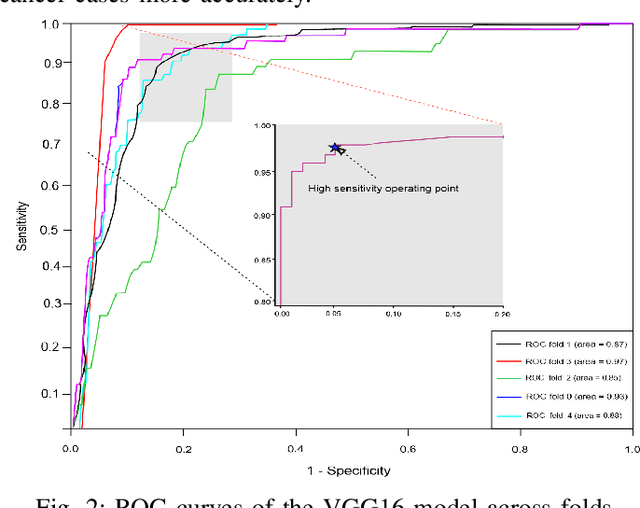
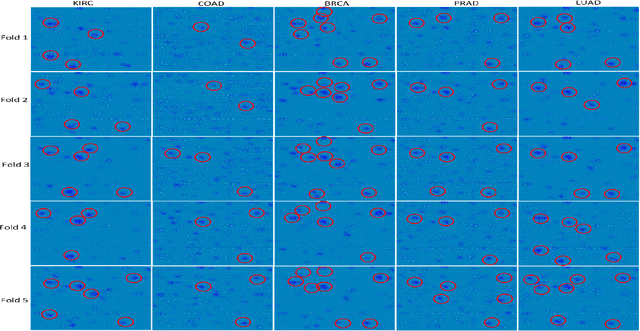
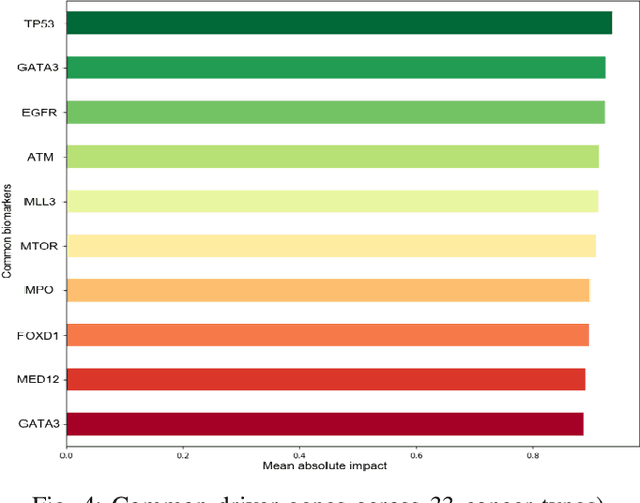
Abstract:The discovery of important biomarkers is a significant step towards understanding the molecular mechanisms of carcinogenesis; enabling accurate diagnosis for, and prognosis of, a certain cancer type. Before recommending any diagnosis, genomics data such as gene expressions(GE) and clinical outcomes need to be analyzed. However, complex nature, high dimensionality, and heterogeneity in genomics data make the overall analysis challenging. Convolutional neural networks(CNN) have shown tremendous success in solving such problems. However, neural network models are perceived mostly as `black box' methods because of their not well-understood internal functioning. However, interpretability is important to provide insights on why a given cancer case has a certain type. Besides, finding the most important biomarkers can help in recommending more accurate treatments and drug repositioning. In this paper, we propose a new approach called OncoNetExplainer to make explainable predictions of cancer types based on GE data. We used genomics data about 9,074 cancer patients covering 33 different cancer types from the Pan-Cancer Atlas on which we trained CNN and VGG16 networks using guided-gradient class activation maps++(GradCAM++). Further, we generate class-specific heat maps to identify significant biomarkers and computed feature importance in terms of mean absolute impact to rank top genes across all the cancer types. Quantitative and qualitative analyses show that both models exhibit high confidence at predicting the cancer types correctly giving an average precision of 96.25%. To provide comparisons with the baselines, we identified top genes, and cancer-specific driver genes using gradient boosted trees and SHapley Additive exPlanations(SHAP). Finally, our findings were validated with the annotations provided by the TumorPortal.
* In proc. of 19th IEEE International Conference on Bioinformatics and Bioengineering(IEEE BIBE 2019)
The Distributed Ontology Language (DOL): Use Cases, Syntax, and Extensibility
Aug 01, 2012
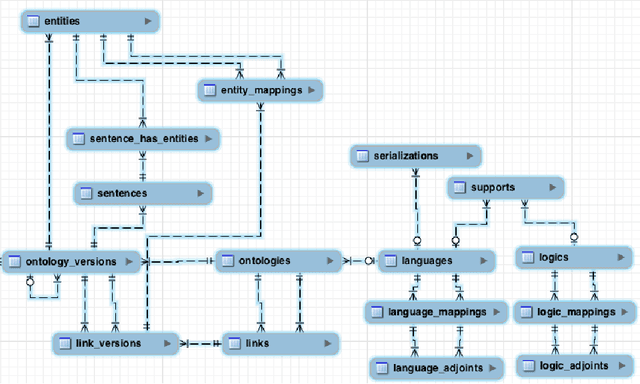
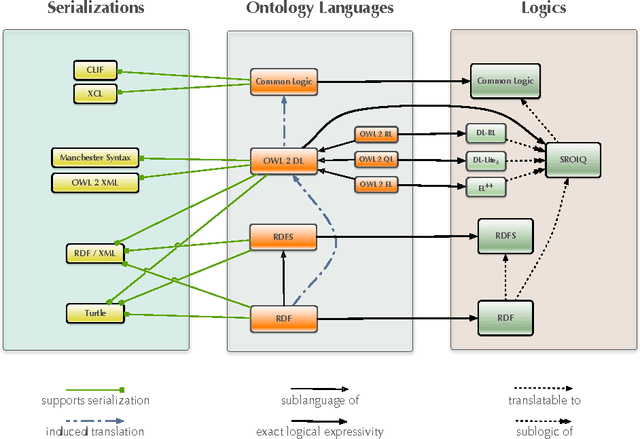
Abstract:The Distributed Ontology Language (DOL) is currently being standardized within the OntoIOp (Ontology Integration and Interoperability) activity of ISO/TC 37/SC 3. It aims at providing a unified framework for (1) ontologies formalized in heterogeneous logics, (2) modular ontologies, (3) links between ontologies, and (4) annotation of ontologies. This paper presents the current state of DOL's standardization. It focuses on use cases where distributed ontologies enable interoperability and reusability. We demonstrate relevant features of the DOL syntax and semantics and explain how these integrate into existing knowledge engineering environments.
sTeX+ - a System for Flexible Formalization of Linked Data
Jun 23, 2010
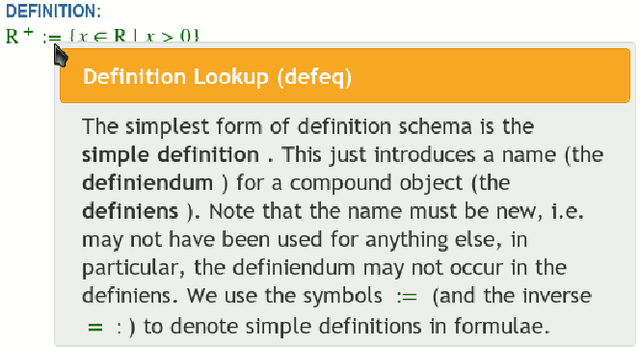
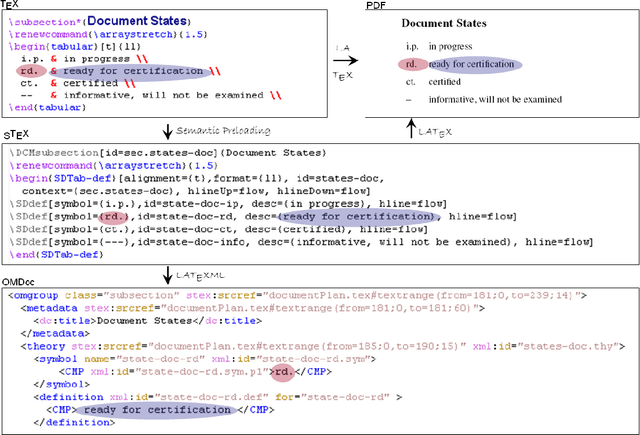
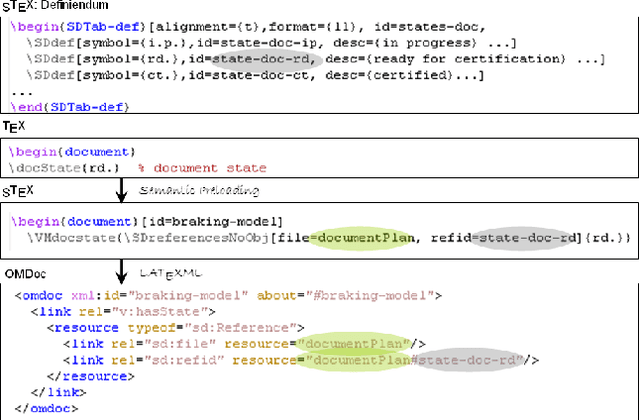
Abstract:We present the sTeX+ system, a user-driven advancement of sTeX - a semantic extension of LaTeX that allows for producing high-quality PDF documents for (proof)reading and printing, as well as semantic XML/OMDoc documents for the Web or further processing. Originally sTeX had been created as an invasive, semantic frontend for authoring XML documents. Here, we used sTeX in a Software Engineering case study as a formalization tool. In order to deal with modular pre-semantic vocabularies and relations, we upgraded it to sTeX+ in a participatory design process. We present a tool chain that starts with an sTeX+ editor and ultimately serves the generated documents as XHTML+RDFa Linked Data via an OMDoc-enabled, versioned XML database. In the final output, all structural annotations are preserved in order to enable semantic information retrieval services.
Dimensions of Formality: A Case Study for MKM in Software Engineering
Apr 28, 2010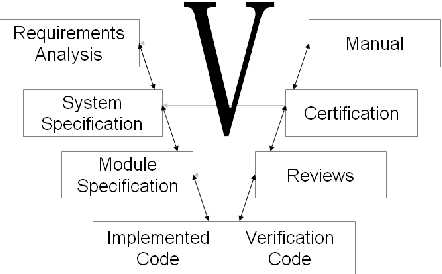

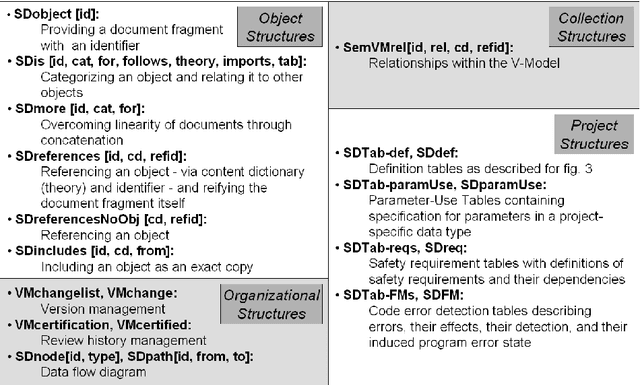

Abstract:We study the formalization of a collection of documents created for a Software Engineering project from an MKM perspective. We analyze how document and collection markup formats can cope with an open-ended, multi-dimensional space of primary and secondary classifications and relationships. We show that RDFa-based extensions of MKM formats, employing flexible "metadata" relationships referencing specific vocabularies for distinct dimensions, are well-suited to encode this and to put it into service. This formalized knowledge can be used for enriching interactive document browsing, for enabling multi-dimensional metadata queries over documents and collections, and for exporting Linked Data to the Semantic Web and thus enabling further reuse.
Publishing Math Lecture Notes as Linked Data
Apr 20, 2010Abstract:We mark up a corpus of LaTeX lecture notes semantically and expose them as Linked Data in XHTML+MathML+RDFa. Our application makes the resulting documents interactively browsable for students. Our ontology helps to answer queries from students and lecturers, and paves the path towards an integration of our corpus with external sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge