Peter Neubauer
Data Augmentation Scheme for Raman Spectra with Highly Correlated Annotations
Feb 01, 2024Abstract:In biotechnology Raman Spectroscopy is rapidly gaining popularity as a process analytical technology (PAT) that measures cell densities, substrate- and product concentrations. As it records vibrational modes of molecules it provides that information non-invasively in a single spectrum. Typically, partial least squares (PLS) is the model of choice to infer information about variables of interest from the spectra. However, biological processes are known for their complexity where convolutional neural networks (CNN) present a powerful alternative. They can handle non-Gaussian noise and account for beam misalignment, pixel malfunctions or the presence of additional substances. However, they require a lot of data during model training, and they pick up non-linear dependencies in the process variables. In this work, we exploit the additive nature of spectra in order to generate additional data points from a given dataset that have statistically independent labels so that a network trained on such data exhibits low correlations between the model predictions. We show that training a CNN on these generated data points improves the performance on datasets where the annotations do not bear the same correlation as the dataset that was used for model training. This data augmentation technique enables us to reuse spectra as training data for new contexts that exhibit different correlations. The additional data allows for building a better and more robust model. This is of interest in scenarios where large amounts of historical data are available but are currently not used for model training. We demonstrate the capabilities of the proposed method using synthetic spectra of Ralstonia eutropha batch cultivations to monitor substrate, biomass and polyhydroxyalkanoate (PHA) biopolymer concentrations during of the experiments.
Deep Set Neural Networks for forecasting asynchronous bioprocess timeseries
Dec 05, 2023Abstract:Cultivation experiments often produce sparse and irregular time series. Classical approaches based on mechanistic models, like Maximum Likelihood fitting or Monte-Carlo Markov chain sampling, can easily account for sparsity and time-grid irregularities, but most statistical and Machine Learning tools are not designed for handling sparse data out-of-the-box. Among popular approaches there are various schemes for filling missing values (imputation) and interpolation into a regular grid (alignment). However, such methods transfer the biases of the interpolation or imputation models to the target model. We show that Deep Set Neural Networks equipped with triplet encoding of the input data can successfully handle bio-process data without any need for imputation or alignment procedures. The method is agnostic to the particular nature of the time series and can be adapted for any task, for example, online monitoring, predictive control, design of experiments, etc. In this work, we focus on forecasting. We argue that such an approach is especially suitable for typical cultivation processes, demonstrate the performance of the method on several forecasting tasks using data generated from macrokinetic growth models under realistic conditions, and compare the method to a conventional fitting procedure and methods based on imputation and alignment.
Deep Learning for Fast Inference of Mechanistic Models' Parameters
Dec 05, 2023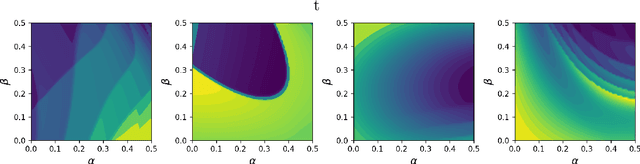
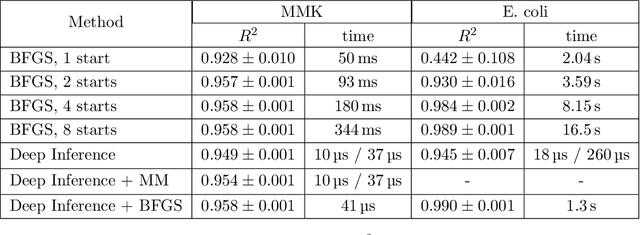
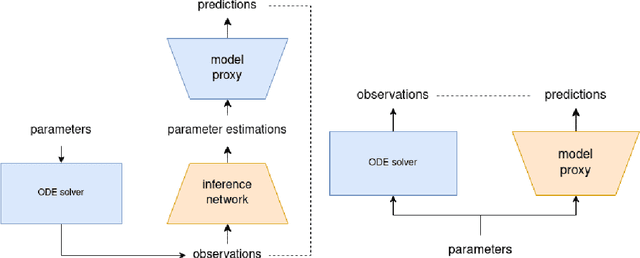
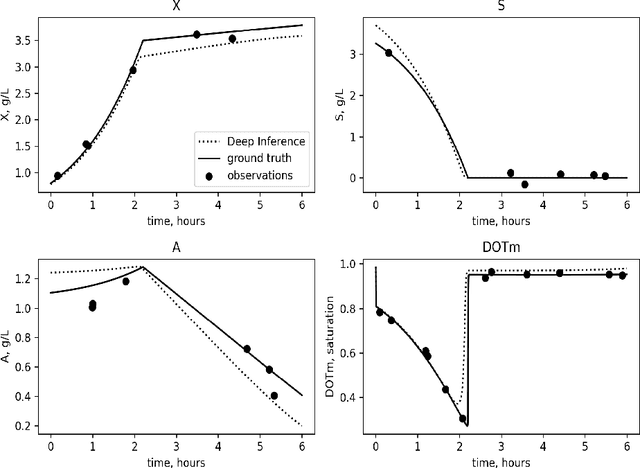
Abstract:Inferring parameters of macro-kinetic growth models, typically represented by Ordinary Differential Equations (ODE), from the experimental data is a crucial step in bioprocess engineering. Conventionally, estimates of the parameters are obtained by fitting the mechanistic model to observations. Fitting, however, requires a significant computational power. Specifically, during the development of new bioprocesses that use previously unknown organisms or strains, efficient, robust, and computationally cheap methods for parameter estimation are of great value. In this work, we propose using Deep Neural Networks (NN) for directly predicting parameters of mechanistic models given observations. The approach requires spending computational resources for training a NN, nonetheless, once trained, such a network can provide parameter estimates orders of magnitude faster than conventional methods. We consider a training procedure that combines Neural Networks and mechanistic models. We demonstrate the performance of the proposed algorithms on data sampled from several mechanistic models used in bioengineering describing a typical industrial batch process and compare the proposed method, a typical gradient-based fitting procedure, and the combination of the two. We find that, while Neural Network estimates are slightly improved by further fitting, these estimates are measurably better than the fitting procedure alone.
When Bioprocess Engineering Meets Machine Learning: A Survey from the Perspective of Automated Bioprocess Development
Sep 02, 2022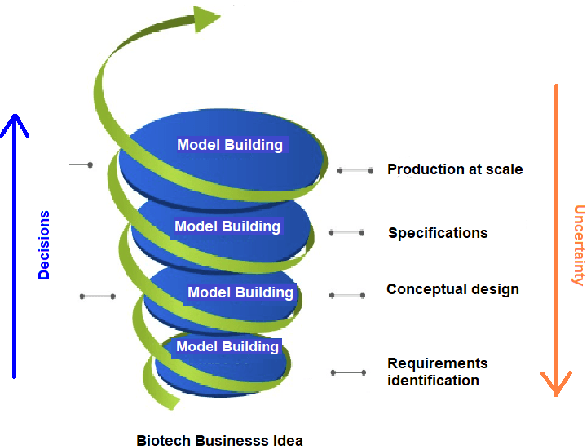
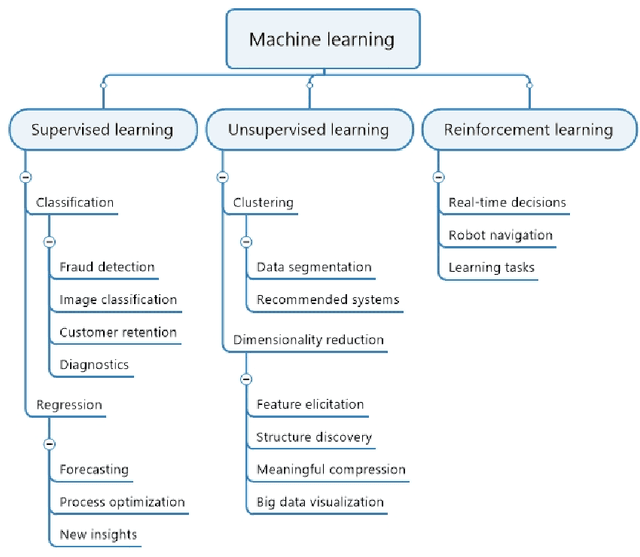
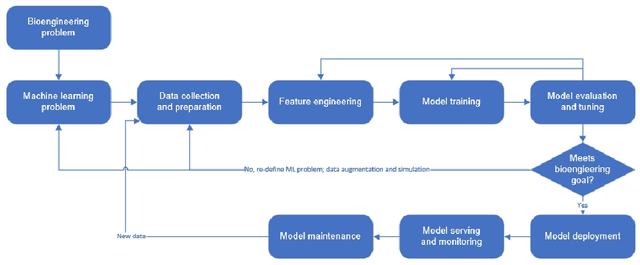
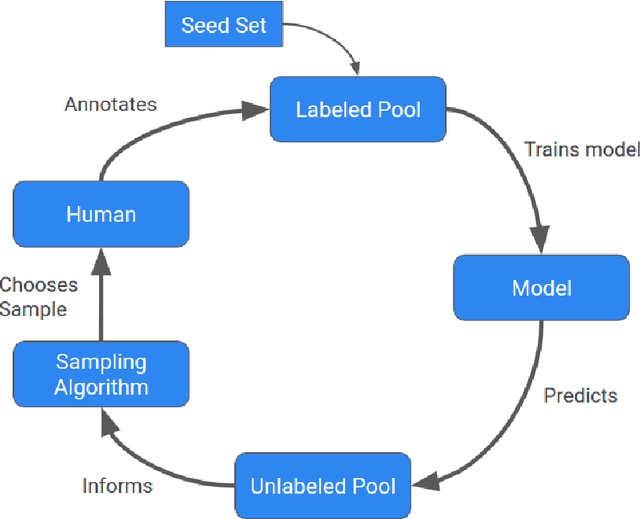
Abstract:Machine learning (ML) has significantly contributed to the development of bioprocess engineering, but its application is still limited, hampering the enormous potential for bioprocess automation. ML for model building automation can be seen as a way of introducing another level of abstraction to focus expert humans in the most cognitive tasks of bioprocess development. First, probabilistic programming is used for the autonomous building of predictive models. Second, machine learning automatically assesses alternative decisions by planning experiments to test hypotheses and conducting investigations to gather informative data that focus on model selection based on the uncertainty of model predictions. This review provides a comprehensive overview of ML-based automation in bioprocess development. On the one hand, the biotech and bioengineering community should be aware of the potential and, most importantly, the limitation of existing ML solutions for their application in biotechnology and biopharma. On the other hand, it is essential to identify the missing links to enable the easy implementation of ML and Artificial Intelligence (AI) solutions in valuable solutions for the bio-community. We summarize recent ML implementation across several important subfields of bioprocess systems and raise two crucial challenges remaining the bottleneck of bioprocess automation and reducing uncertainty in biotechnology development. There is no one-fits-all procedure; however, this review should help identify the potential automation combining biotechnology and ML domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge