Thorben Werner
The Role of Active Learning in Modern Machine Learning
Aug 01, 2025Abstract:Even though Active Learning (AL) is widely studied, it is rarely applied in contexts outside its own scientific literature. We posit that the reason for this is AL's high computational cost coupled with the comparatively small lifts it is typically able to generate in scenarios with few labeled points. In this work we study the impact of different methods to combat this low data scenario, namely data augmentation (DA), semi-supervised learning (SSL) and AL. We find that AL is by far the least efficient method of solving the low data problem, generating a lift of only 1-4\% over random sampling, while DA and SSL methods can generate up to 60\% lift in combination with random sampling. However, when AL is combined with strong DA and SSL techniques, it surprisingly is still able to provide improvements. Based on these results, we frame AL not as a method to combat missing labels, but as the final building block to squeeze the last bits of performance out of data after appropriate DA and SSL methods as been applied.
Bayesian Active Learning By Distribution Disagreement
Jan 02, 2025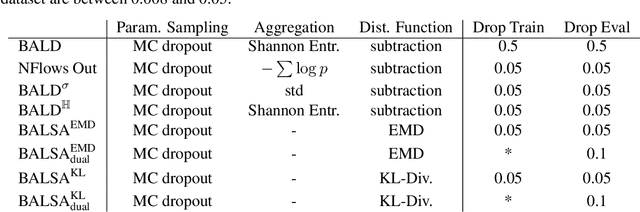
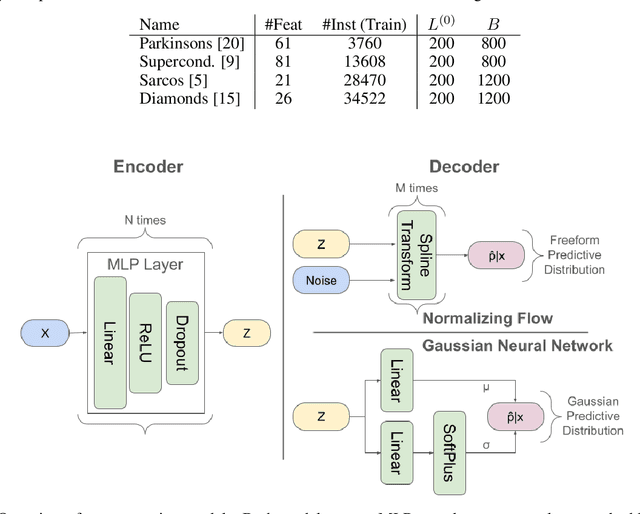
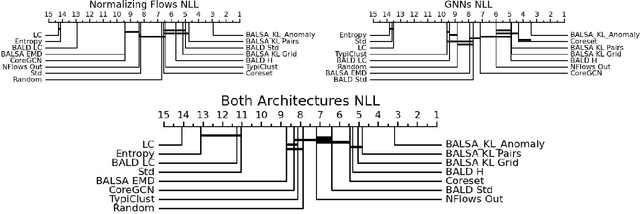
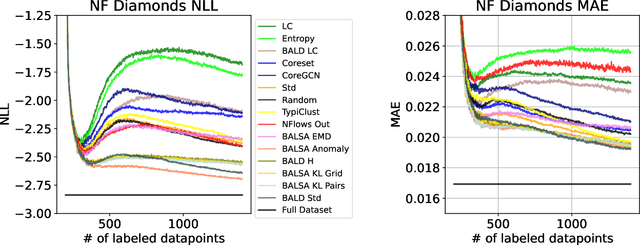
Abstract:Active Learning (AL) for regression has been systematically under-researched due to the increased difficulty of measuring uncertainty in regression models. Since normalizing flows offer a full predictive distribution instead of a point forecast, they facilitate direct usage of known heuristics for AL like Entropy or Least-Confident sampling. However, we show that most of these heuristics do not work well for normalizing flows in pool-based AL and we need more sophisticated algorithms to distinguish between aleatoric and epistemic uncertainty. In this work we propose BALSA, an adaptation of the BALD algorithm, tailored for regression with normalizing flows. With this work we extend current research on uncertainty quantification with normalizing flows \cite{berry2023normalizing, berry2023escaping} to real world data and pool-based AL with multiple acquisition functions and query sizes. We report SOTA results for BALSA across 4 different datasets and 2 different architectures.
A Cross-Domain Benchmark for Active Learning
Aug 01, 2024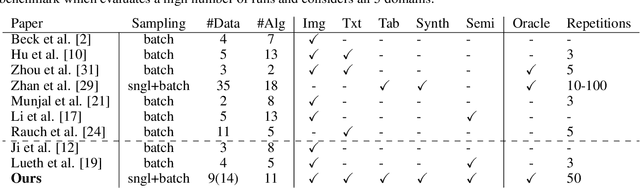
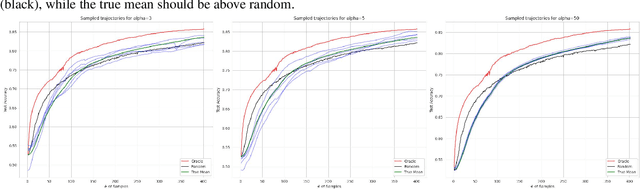
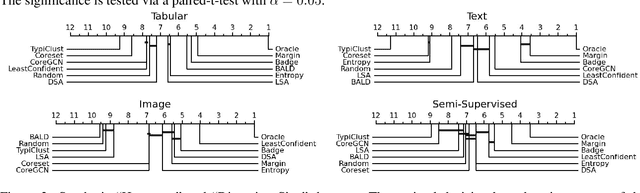
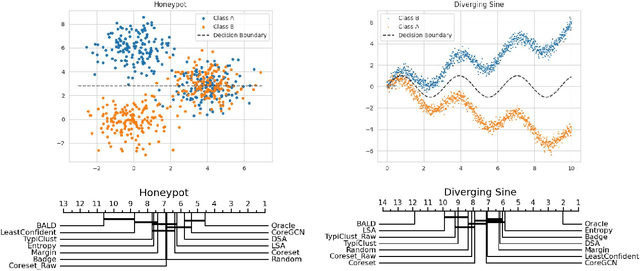
Abstract:Active Learning (AL) deals with identifying the most informative samples for labeling to reduce data annotation costs for supervised learning tasks. AL research suffers from the fact that lifts from literature generalize poorly and that only a small number of repetitions of experiments are conducted. To overcome these obstacles, we propose \emph{CDALBench}, the first active learning benchmark which includes tasks in computer vision, natural language processing and tabular learning. Furthermore, by providing an efficient, greedy oracle, \emph{CDALBench} can be evaluated with 50 runs for each experiment. We show, that both the cross-domain character and a large amount of repetitions are crucial for sophisticated evaluation of AL research. Concretely, we show that the superiority of specific methods varies over the different domains, making it important to evaluate Active Learning with a cross-domain benchmark. Additionally, we show that having a large amount of runs is crucial. With only conducting three runs as often done in the literature, the superiority of specific methods can strongly vary with the specific runs. This effect is so strong, that, depending on the seed, even a well-established method's performance can be significantly better and significantly worse than random for the same dataset.
Are EEG Sequences Time Series? EEG Classification with Time Series Models and Joint Subject Training
Apr 10, 2024



Abstract:As with most other data domains, EEG data analysis relies on rich domain-specific preprocessing. Beyond such preprocessing, machine learners would hope to deal with such data as with any other time series data. For EEG classification many models have been developed with layer types and architectures we typically do not see in time series classification. Furthermore, typically separate models for each individual subject are learned, not one model for all of them. In this paper, we systematically study the differences between EEG classification models and generic time series classification models. We describe three different model setups to deal with EEG data from different subjects, subject-specific models (most EEG literature), subject-agnostic models and subject-conditional models. In experiments on three datasets, we demonstrate that off-the-shelf time series classification models trained per subject perform close to EEG classification models, but that do not quite reach the performance of domain-specific modeling. Additionally, we combine time-series models with subject embeddings to train one joint subject-conditional classifier on all subjects. The resulting models are competitive with dedicated EEG models in 2 out of 3 datasets, even outperforming all EEG methods on one of them.
Towards Comparable Active Learning
Nov 30, 2023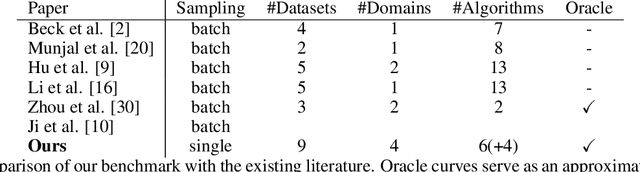
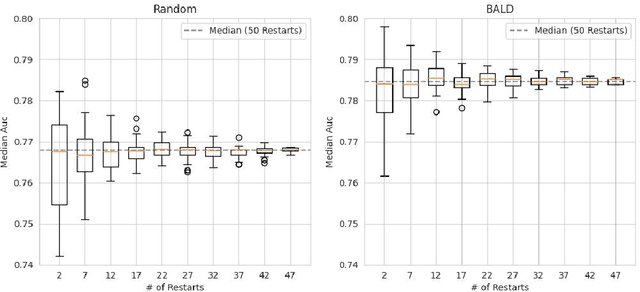
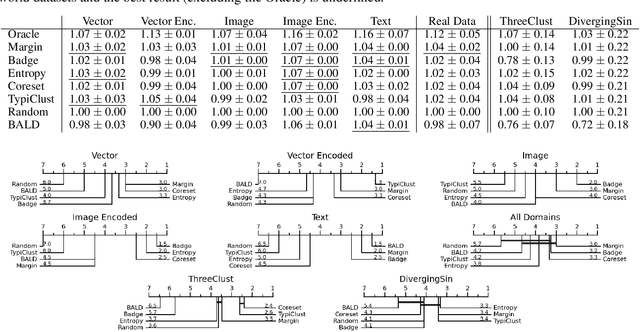
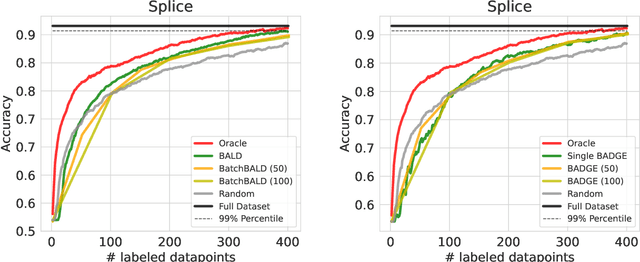
Abstract:Active Learning has received significant attention in the field of machine learning for its potential in selecting the most informative samples for labeling, thereby reducing data annotation costs. However, we show that the reported lifts in recent literature generalize poorly to other domains leading to an inconclusive landscape in Active Learning research. Furthermore, we highlight overlooked problems for reproducing AL experiments that can lead to unfair comparisons and increased variance in the results. This paper addresses these issues by providing an Active Learning framework for a fair comparison of algorithms across different tasks and domains, as well as a fast and performant oracle algorithm for evaluation. To the best of our knowledge, we propose the first AL benchmark that tests algorithms in 3 major domains: Tabular, Image, and Text. We report empirical results for 6 widely used algorithms on 7 real-world and 2 synthetic datasets and aggregate them into a domain-specific ranking of AL algorithms.
Towards Comparable Knowledge Distillation in Semantic Image Segmentation
Sep 07, 2023Abstract:Knowledge Distillation (KD) is one proposed solution to large model sizes and slow inference speed in semantic segmentation. In our research we identify 25 proposed distillation loss terms from 14 publications in the last 4 years. Unfortunately, a comparison of terms based on published results is often impossible, because of differences in training configurations. A good illustration of this problem is the comparison of two publications from 2022. Using the same models and dataset, Structural and Statistical Texture Distillation (SSTKD) reports an increase of student mIoU of 4.54 and a final performance of 29.19, while Adaptive Perspective Distillation (APD) only improves student performance by 2.06 percentage points, but achieves a final performance of 39.25. The reason for such extreme differences is often a suboptimal choice of hyperparameters and a resulting underperformance of the student model used as reference point. In our work, we reveal problems of insufficient hyperparameter tuning by showing that distillation improvements of two widely accepted frameworks, SKD and IFVD, vanish when hyperparameters are optimized sufficiently. To improve comparability of future research in the field, we establish a solid baseline for three datasets and two student models and provide extensive information on hyperparameter tuning. We find that only two out of eight techniques can compete with our simple baseline on the ADE20K dataset.
When Bioprocess Engineering Meets Machine Learning: A Survey from the Perspective of Automated Bioprocess Development
Sep 02, 2022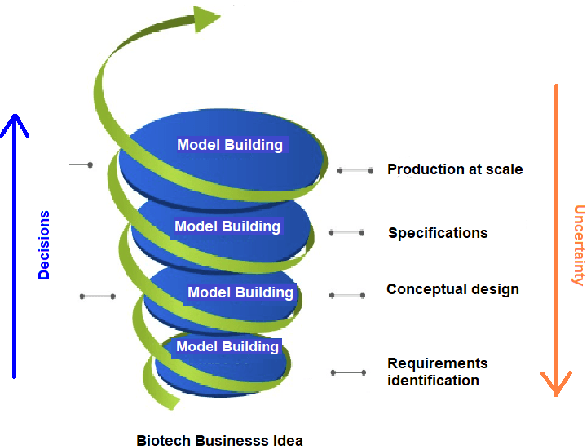
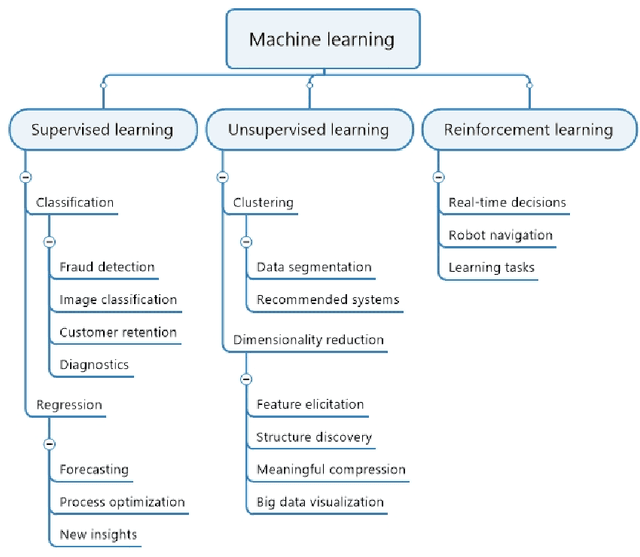
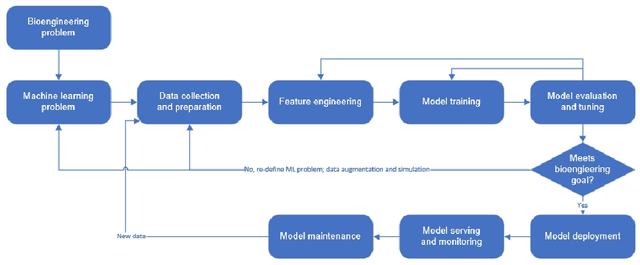
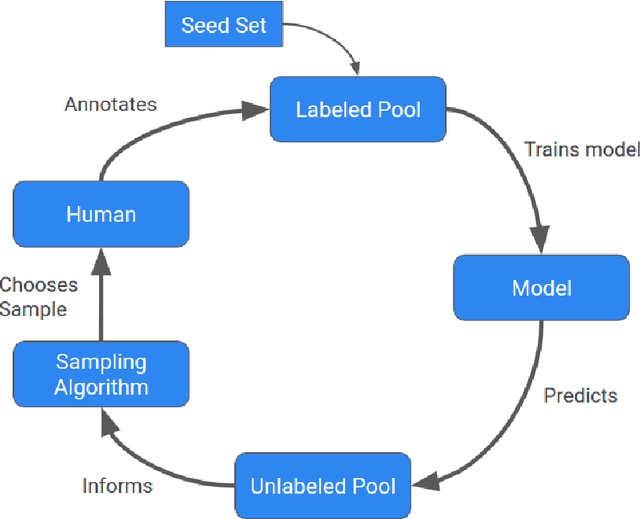
Abstract:Machine learning (ML) has significantly contributed to the development of bioprocess engineering, but its application is still limited, hampering the enormous potential for bioprocess automation. ML for model building automation can be seen as a way of introducing another level of abstraction to focus expert humans in the most cognitive tasks of bioprocess development. First, probabilistic programming is used for the autonomous building of predictive models. Second, machine learning automatically assesses alternative decisions by planning experiments to test hypotheses and conducting investigations to gather informative data that focus on model selection based on the uncertainty of model predictions. This review provides a comprehensive overview of ML-based automation in bioprocess development. On the one hand, the biotech and bioengineering community should be aware of the potential and, most importantly, the limitation of existing ML solutions for their application in biotechnology and biopharma. On the other hand, it is essential to identify the missing links to enable the easy implementation of ML and Artificial Intelligence (AI) solutions in valuable solutions for the bio-community. We summarize recent ML implementation across several important subfields of bioprocess systems and raise two crucial challenges remaining the bottleneck of bioprocess automation and reducing uncertainty in biotechnology development. There is no one-fits-all procedure; however, this review should help identify the potential automation combining biotechnology and ML domains.
Reinforcement Learning Approach to Active Learning for Image Classification
Aug 12, 2021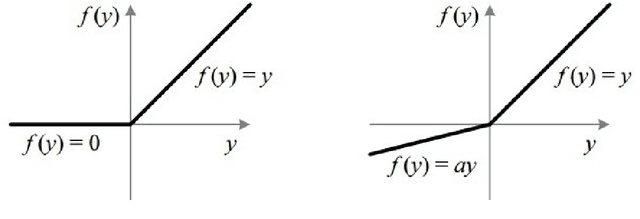
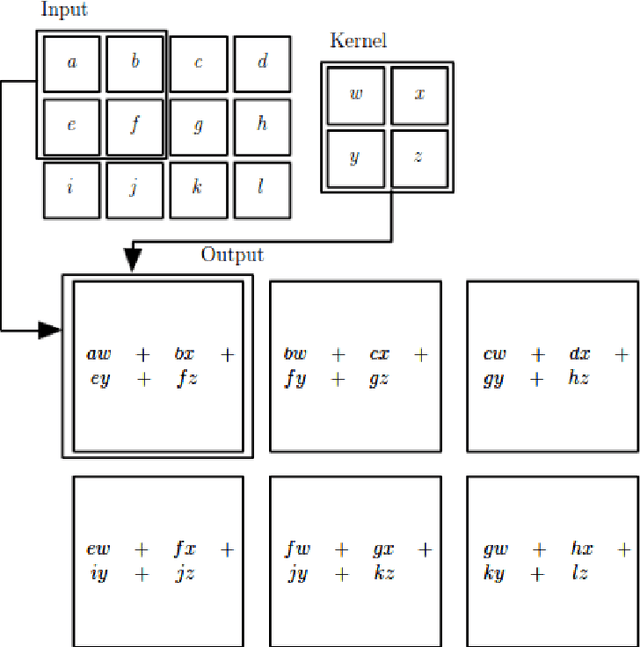
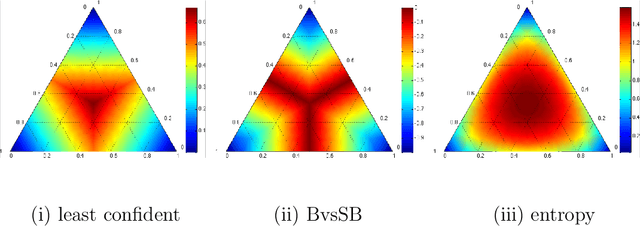
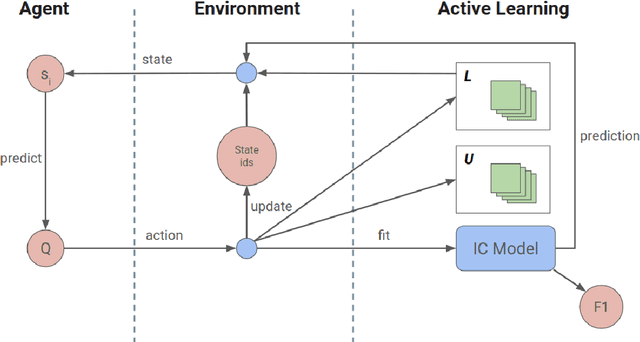
Abstract:Machine Learning requires large amounts of labeled data to fit a model. Many datasets are already publicly available, nevertheless forcing application possibilities of machine learning to the domains of those public datasets. The ever-growing penetration of machine learning algorithms in new application areas requires solutions for the need for data in those new domains. This thesis works on active learning as one possible solution to reduce the amount of data that needs to be processed by hand, by processing only those datapoints that specifically benefit the training of a strong model for the task. A newly proposed framework for framing the active learning workflow as a reinforcement learning problem is adapted for image classification and a series of three experiments is conducted. Each experiment is evaluated and potential issues with the approach are outlined. Each following experiment then proposes improvements to the framework and evaluates their impact. After the last experiment, a final conclusion is drawn, unfortunately rejecting this work's hypothesis and outlining that the proposed framework at the moment is not capable of improving active learning for image classification with a trained reinforcement learning agent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge