A Biomedical Knowledge Graph for Biomarker Discovery in Cancer
Paper and Code
Feb 23, 2023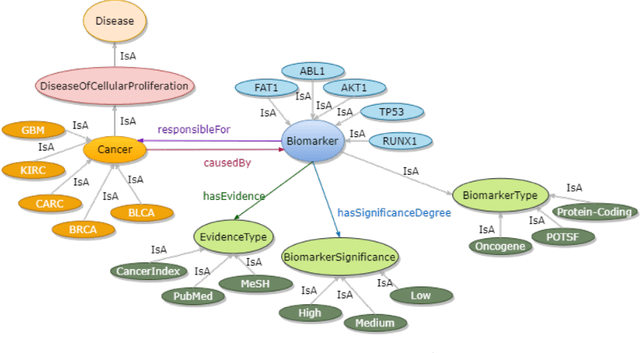
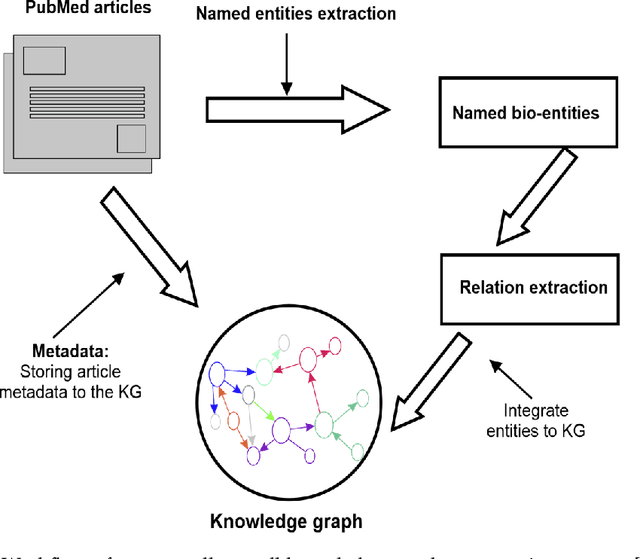

Structured and unstructured data and facts about drugs, genes, protein, viruses, and their mechanism are spread across a huge number of scientific articles. These articles are a large-scale knowledge source and can have a huge impact on disseminating knowledge about the mechanisms of certain biological processes. A domain-specific knowledge graph~(KG) is an explicit conceptualization of a specific subject-matter domain represented w.r.t semantically interrelated entities and relations. A KG can be constructed by integrating such facts and data and be used for data integration, exploration, and federated queries. However, exploration and querying large-scale KGs is tedious for certain groups of users due to a lack of knowledge about underlying data assets or semantic technologies. Such a KG will not only allow deducing new knowledge and question answering(QA) but also allows domain experts to explore. Since cross-disciplinary explanations are important for accurate diagnosis, it is important to query the KG to provide interactive explanations about learned biomarkers. Inspired by these, we construct a domain-specific KG, particularly for cancer-specific biomarker discovery. The KG is constructed by integrating cancer-related knowledge and facts from multiple sources. First, we construct a domain-specific ontology, which we call OncoNet Ontology (ONO). The ONO ontology is developed to enable semantic reasoning for verification of the predictions for relations between diseases and genes. The KG is then developed and enriched by harmonizing the ONO, additional metadata schemas, ontologies, controlled vocabularies, and additional concepts from external sources using a BERT-based information extraction method. BioBERT and SciBERT are finetuned with the selected articles crawled from PubMed. We listed down some queries and some examples of QA and deducing knowledge based on the KG.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge