Shunjie Dong
SFCNeXt: a simple fully convolutional network for effective brain age estimation with small sample size
May 30, 2023Abstract:Deep neural networks (DNN) have been designed to predict the chronological age of a healthy brain from T1-weighted magnetic resonance images (T1 MRIs), and the predicted brain age could serve as a valuable biomarker for the early detection of development-related or aging-related disorders. Recent DNN models for brain age estimations usually rely too much on large sample sizes and complex network structures for multi-stage feature refinement. However, in clinical application scenarios, researchers usually cannot obtain thousands or tens of thousands of MRIs in each data center for thorough training of these complex models. This paper proposes a simple fully convolutional network (SFCNeXt) for brain age estimation in small-sized cohorts with biased age distributions. The SFCNeXt consists of Single Pathway Encoded ConvNeXt (SPEC) and Hybrid Ranking Loss (HRL), aiming to estimate brain ages in a lightweight way with a sufficient exploration of MRI, age, and ranking features of each batch of subjects. Experimental results demonstrate the superiority and efficiency of our approach.
RT-DNAS: Real-time Constrained Differentiable Neural Architecture Search for 3D Cardiac Cine MRI Segmentation
Jun 13, 2022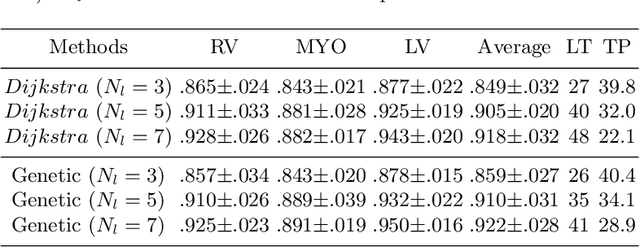
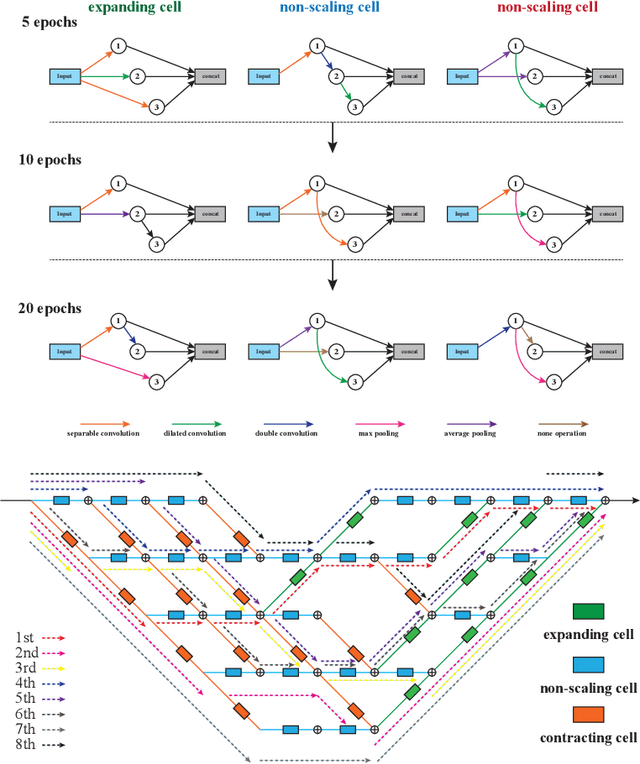
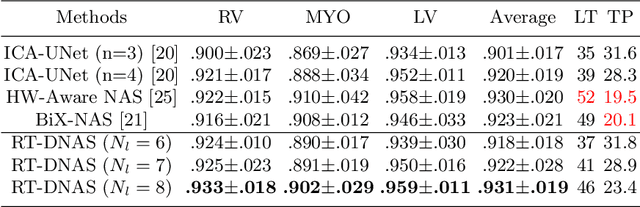
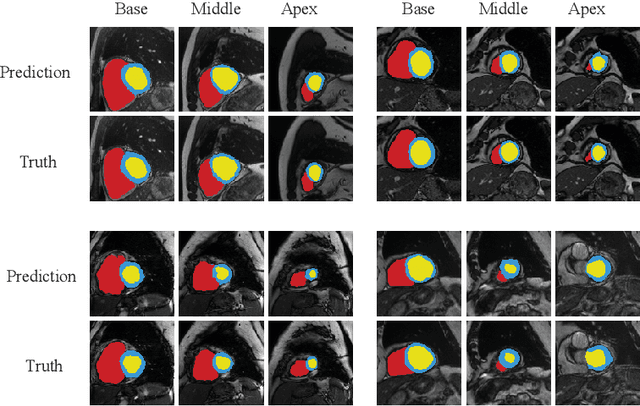
Abstract:Accurately segmenting temporal frames of cine magnetic resonance imaging (MRI) is a crucial step in various real-time MRI guided cardiac interventions. To achieve fast and accurate visual assistance, there are strict requirements on the maximum latency and minimum throughput of the segmentation framework. State-of-the-art neural networks on this task are mostly hand-crafted to satisfy these constraints while achieving high accuracy. On the other hand, while existing literature have demonstrated the power of neural architecture search (NAS) in automatically identifying the best neural architectures for various medical applications, they are mostly guided by accuracy, sometimes with computation complexity, and the importance of real-time constraints are overlooked. A major challenge is that such constraints are non-differentiable and are thus not compatible with the widely used differentiable NAS frameworks. In this paper, we present a strategy that directly handles real-time constraints in a differentiable NAS framework named RT-DNAS. Experiments on extended 2017 MICCAI ACDC dataset show that compared with state-of-the-art manually and automatically designed architectures, RT-DNAS is able to identify ones with better accuracy while satisfying the real-time constraints.
OTFPF: Optimal Transport-Based Feature Pyramid Fusion Network for Brain Age Estimation with 3D Overlapped ConvNeXt
May 11, 2022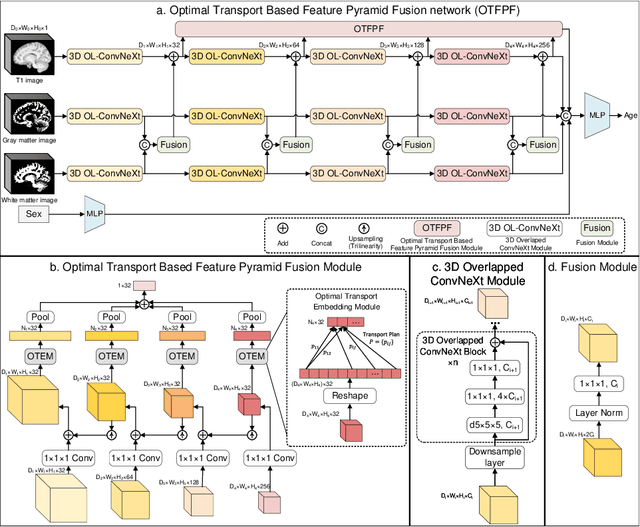
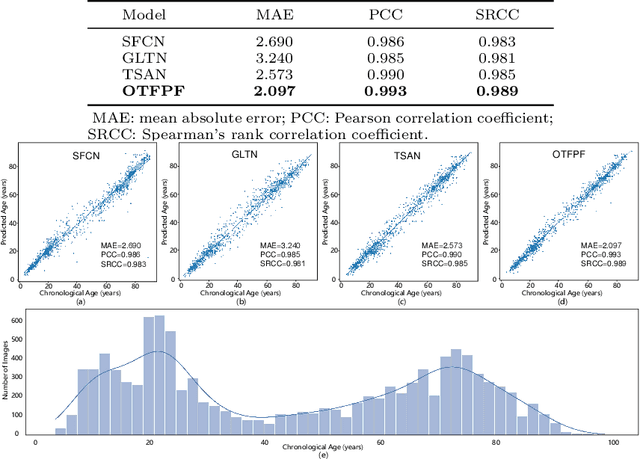
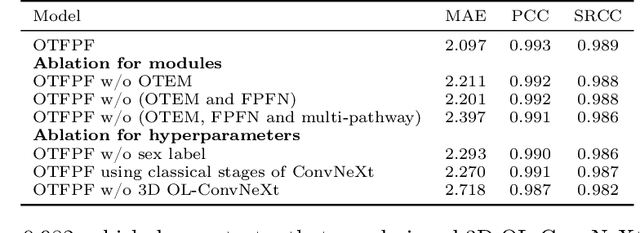
Abstract:Chronological age of healthy brain is able to be predicted using deep neural networks from T1-weighted magnetic resonance images (T1 MRIs), and the predicted brain age could serve as an effective biomarker for detecting aging-related diseases or disorders. In this paper, we propose an end-to-end neural network architecture, referred to as optimal transport based feature pyramid fusion (OTFPF) network, for the brain age estimation with T1 MRIs. The OTFPF consists of three types of modules: Optimal Transport based Feature Pyramid Fusion (OTFPF) module, 3D overlapped ConvNeXt (3D OL-ConvNeXt) module and fusion module. These modules strengthen the OTFPF network's understanding of each brain's semi-multimodal and multi-level feature pyramid information, and significantly improve its estimation performances. Comparing with recent state-of-the-art models, the proposed OTFPF converges faster and performs better. The experiments with 11,728 MRIs aged 3-97 years show that OTFPF network could provide accurate brain age estimation, yielding mean absolute error (MAE) of 2.097, Pearson's correlation coefficient (PCC) of 0.993 and Spearman's rank correlation coefficient (SRCC) of 0.989, between the estimated and chronological ages. Widespread quantitative experiments and ablation experiments demonstrate the superiority and rationality of OTFPF network. The codes and implement details will be released on GitHub: https://github.com/ZJU-Brain/OTFPF after final decision.
A resource-efficient deep learning framework for low-dose brain PET image reconstruction and analysis
Feb 14, 2022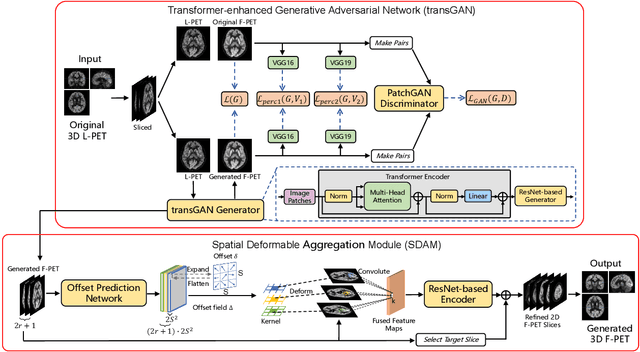

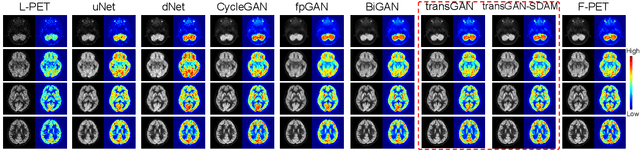
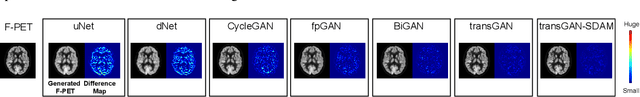
Abstract:18F-fluorodeoxyglucose (18F-FDG) Positron Emission Tomography (PET) imaging usually needs a full-dose radioactive tracer to obtain satisfactory diagnostic results, which raises concerns about the potential health risks of radiation exposure, especially for pediatric patients. Reconstructing the low-dose PET (L-PET) images to the high-quality full-dose PET (F-PET) ones is an effective way that both reduces the radiation exposure and remains diagnostic accuracy. In this paper, we propose a resource-efficient deep learning framework for L-PET reconstruction and analysis, referred to as transGAN-SDAM, to generate F-PET from corresponding L-PET, and quantify the standard uptake value ratios (SUVRs) of these generated F-PET at whole brain. The transGAN-SDAM consists of two modules: a transformer-encoded Generative Adversarial Network (transGAN) and a Spatial Deformable Aggregation Module (SDAM). The transGAN generates higher quality F-PET images, and then the SDAM integrates the spatial information of a sequence of generated F-PET slices to synthesize whole-brain F-PET images. Experimental results demonstrate the superiority and rationality of our approach.
RCoNet: Deformable Mutual Information Maximization and High-order Uncertainty-aware Learning for Robust COVID-19 Detection
Feb 22, 2021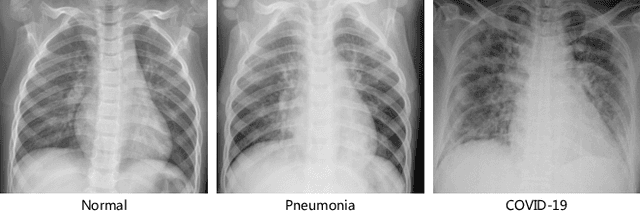
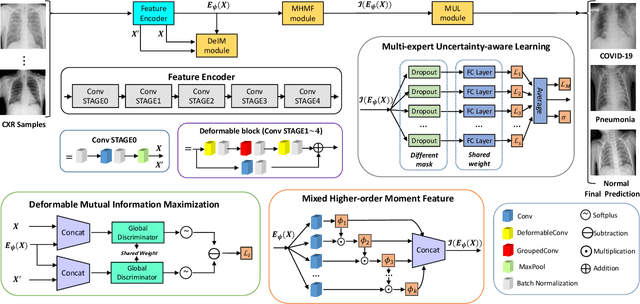
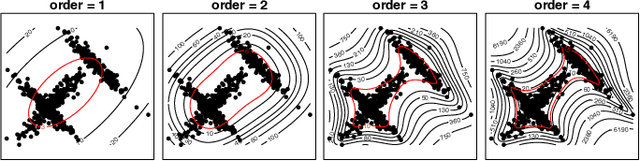
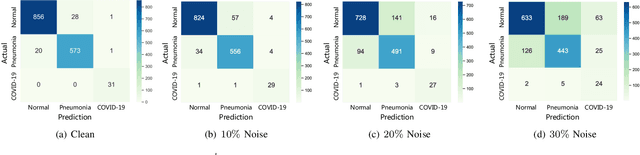
Abstract:The novel 2019 Coronavirus (COVID-19) infection has spread world widely and is currently a major healthcare challenge around the world. Chest Computed Tomography (CT) and X-ray images have been well recognized to be two effective techniques for clinical COVID-19 disease diagnoses. Due to faster imaging time and considerably lower cost than CT, detecting COVID-19 in chest X-ray (CXR) images is preferred for efficient diagnosis, assessment and treatment. However, considering the similarity between COVID-19 and pneumonia, CXR samples with deep features distributed near category boundaries are easily misclassified by the hyper-planes learned from limited training data. Moreover, most existing approaches for COVID-19 detection focus on the accuracy of prediction and overlook the uncertainty estimation, which is particularly important when dealing with noisy datasets. To alleviate these concerns, we propose a novel deep network named {\em RCoNet$^k_s$} for robust COVID-19 detection which employs {\em Deformable Mutual Information Maximization} (DeIM), {\em Mixed High-order Moment Feature} (MHMF) and {\em Multi-expert Uncertainty-aware Learning} (MUL). With DeIM, the mutual information (MI) between input data and the corresponding latent representations can be well estimated and maximized to capture compact and disentangled representational characteristics. Meanwhile, MHMF can fully explore the benefits of using high-order statistics and extract discriminative features of complex distributions in medical imaging. Finally, MUL creates multiple parallel dropout networks for each CXR image to evaluate uncertainty and thus prevent performance degradation caused by the noise in the data.
DeU-Net: Deformable U-Net for 3D Cardiac MRI Video Segmentation
Jul 13, 2020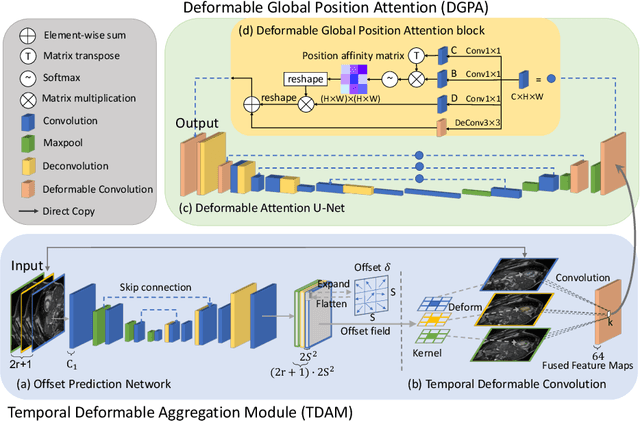
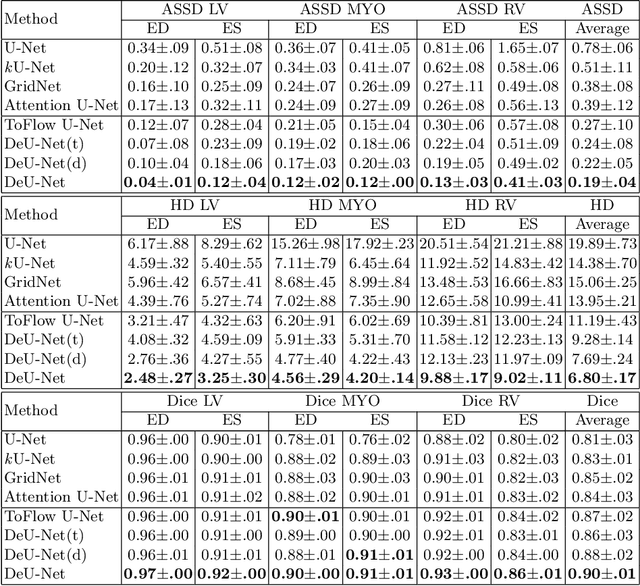
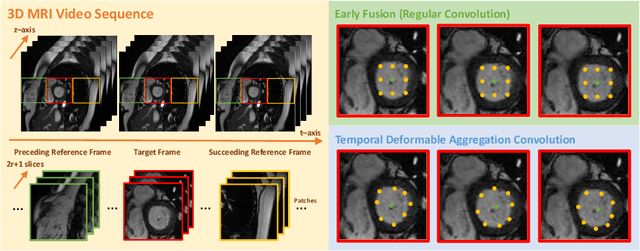
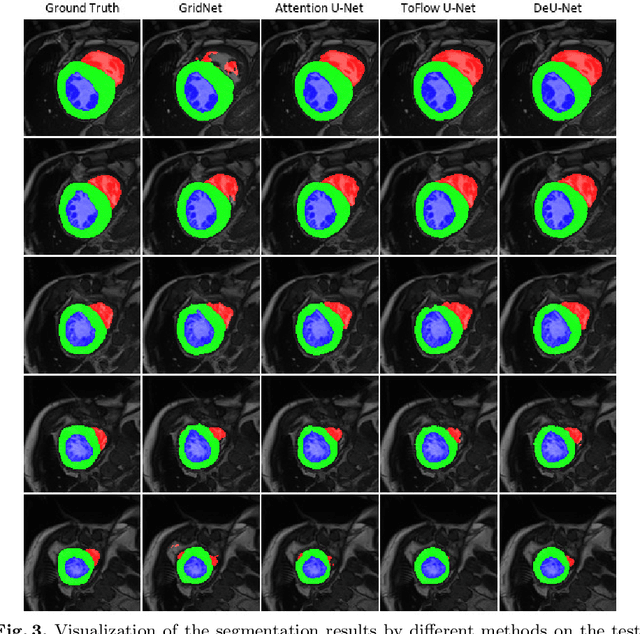
Abstract:Automatic segmentation of cardiac magnetic resonance imaging (MRI) facilitates efficient and accurate volume measurement in clinical applications. However, due to anisotropic resolution and ambiguous border (e.g., right ventricular endocardium), existing methods suffer from the degradation of accuracy and robustness in 3D cardiac MRI video segmentation. In this paper, we propose a novel Deformable U-Net (DeU-Net) to fully exploit spatio-temporal information from 3D cardiac MRI video, including a Temporal Deformable Aggregation Module (TDAM) and a Deformable Global Position Attention (DGPA) network. First, the TDAM takes a cardiac MRI video clip as input with temporal information extracted by an offset prediction network. Then we fuse extracted temporal information via a temporal aggregation deformable convolution to produce fused feature maps. Furthermore, to aggregate meaningful features, we devise the DGPA network by employing deformable attention U-Net, which can encode a wider range of multi-dimensional contextual information into global and local features. Experimental results show that our DeU-Net achieves the state-of-the-art performance on commonly used evaluation metrics, especially for cardiac marginal information (ASSD and HD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge