Niranjan Balachandar
An Experimental Study of Data Heterogeneity in Federated Learning Methods for Medical Imaging
Jul 18, 2021



Abstract:Federated learning enables multiple institutions to collaboratively train machine learning models on their local data in a privacy-preserving way. However, its distributed nature often leads to significant heterogeneity in data distributions across institutions. In this paper, we investigate the deleterious impact of a taxonomy of data heterogeneity regimes on federated learning methods, including quantity skew, label distribution skew, and imaging acquisition skew. We show that the performance degrades with the increasing degrees of data heterogeneity. We present several mitigation strategies to overcome performance drops from data heterogeneity, including weighted average for data quantity skew, weighted loss and batch normalization averaging for label distribution skew. The proposed optimizations to federated learning methods improve their capability of handling heterogeneity across institutions, which provides valuable guidance for the deployment of federated learning in real clinical applications.
Handling Data Heterogeneity with Generative Replay in Collaborative Learning for Medical Imaging
Jun 24, 2021



Abstract:Collaborative learning, which enables collaborative and decentralized training of deep neural networks at multiple institutions in a privacy-preserving manner, is rapidly emerging as a valuable technique in healthcare applications. However, its distributed nature often leads to significant heterogeneity in data distributions across institutions. Existing collaborative learning approaches generally do not account for the presence of heterogeneity in data among institutions, or only mildly skewed label distributions are studied. In this paper, we present a novel generative replay strategy to address the challenge of data heterogeneity in collaborative learning methods. Instead of directly training a model for task performance, we leverage recent image synthesis techniques to develop a novel dual model architecture: a primary model learns the desired task, and an auxiliary "generative replay model" either synthesizes images that closely resemble the input images or helps extract latent variables. The generative replay strategy is flexible to use, can either be incorporated into existing collaborative learning methods to improve their capability of handling data heterogeneity across institutions, or be used as a novel and individual collaborative learning framework (termed FedReplay) to reduce communication cost. Experimental results demonstrate the capability of the proposed method in handling heterogeneous data across institutions. On highly heterogeneous data partitions, our model achieves ~4.88% improvement in the prediction accuracy on a diabetic retinopathy classification dataset, and ~49.8% reduction of mean absolution value on a Bone Age prediction dataset, respectively, compared to the state-of-the art collaborative learning methods.
MedAug: Contrastive learning leveraging patient metadata improves representations for chest X-ray interpretation
Feb 21, 2021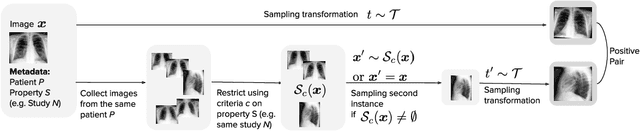
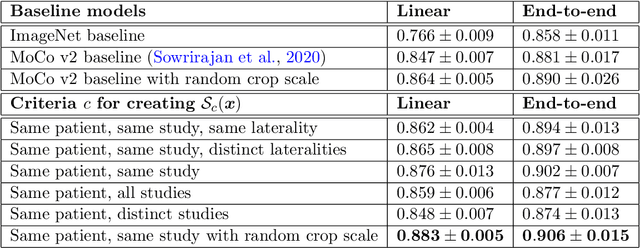

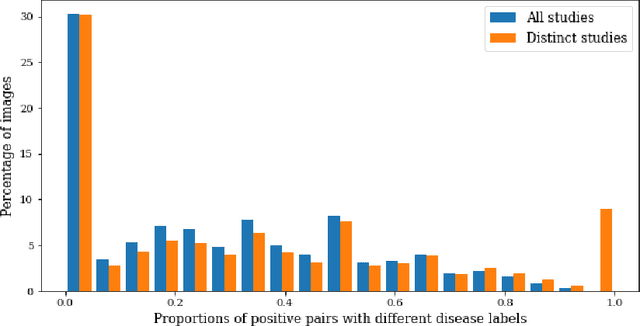
Abstract:Self-supervised contrastive learning between pairs of multiple views of the same image has been shown to successfully leverage unlabeled data to produce meaningful visual representations for both natural and medical images. However, there has been limited work on determining how to select pairs for medical images, where availability of patient metadata can be leveraged to improve representations. In this work, we develop a method to select positive pairs coming from views of possibly different images through the use of patient metadata. We compare strategies for selecting positive pairs for chest X-ray interpretation including requiring them to be from the same patient, imaging study or laterality. We evaluate downstream task performance by fine-tuning the linear layer on 1% of the labeled dataset for pleural effusion classification. Our best performing positive pair selection strategy, which involves using images from the same patient from the same study across all lateralities, achieves a performance increase of 3.4% and 14.4% in mean AUC from both a previous contrastive method and ImageNet pretrained baseline respectively. Our controlled experiments show that the keys to improving downstream performance on disease classification are (1) using patient metadata to appropriately create positive pairs from different images with the same underlying pathologies, and (2) maximizing the number of different images used in query pairing. In addition, we explore leveraging patient metadata to select hard negative pairs for contrastive learning, but do not find improvement over baselines that do not use metadata. Our method is broadly applicable to medical image interpretation and allows flexibility for incorporating medical insights in choosing pairs for contrastive learning.
Prediction of Small Molecule Kinase Inhibitors for Chemotherapy Using Deep Learning
Jun 30, 2019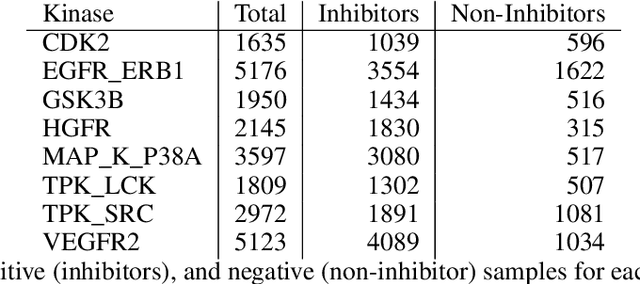
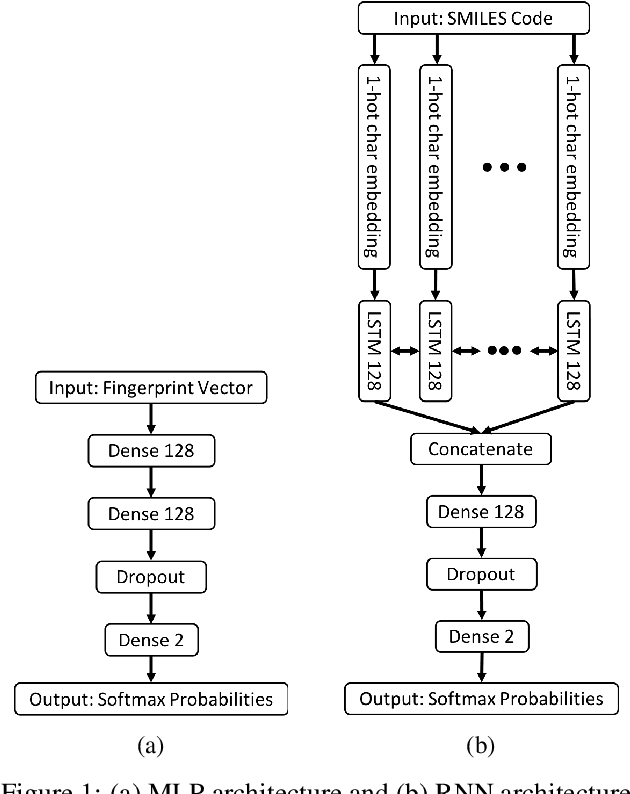
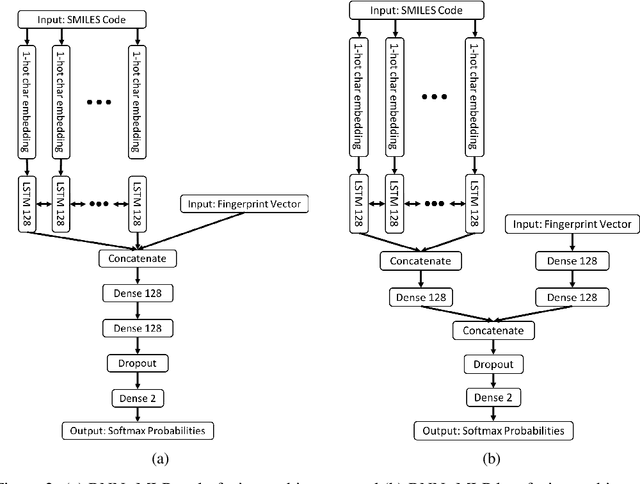
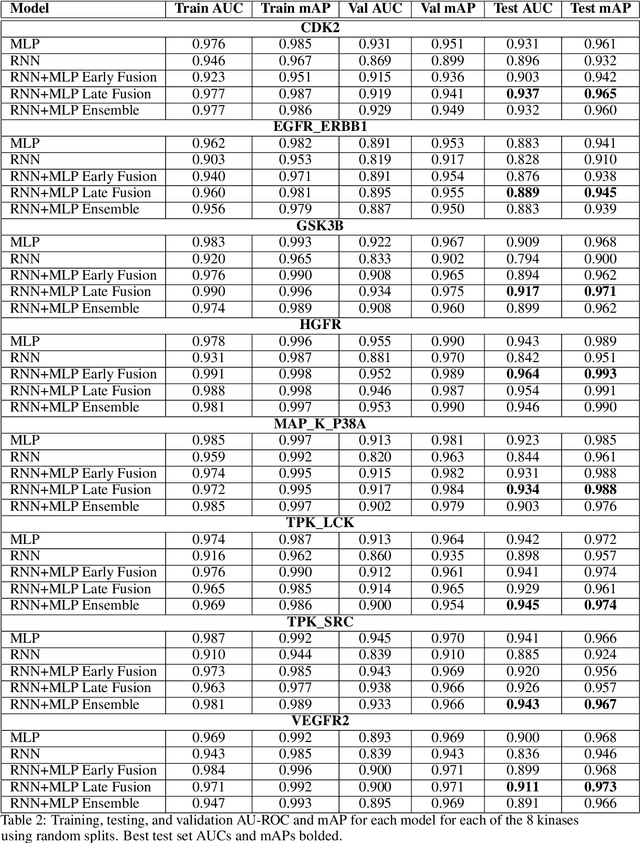
Abstract:The current state of cancer therapeutics has been moving away from one-size-fits-all cytotoxic chemotherapy, and towards a more individualized and specific approach involving the targeting of each tumor's genetic vulnerabilities. Different tumors, even of the same type, may be more reliant on certain cellular pathways more than others. With modern advancements in our understanding of cancer genome sequencing, these pathways can be discovered. Investigating each of the millions of possible small molecule inhibitors for each kinase in vitro, however, would be extremely expensive and time consuming. This project focuses on predicting the inhibition activity of small molecules targeting 8 different kinases using multiple deep learning models. We trained fingerprint-based MLPs and simplified molecular-input line-entry specification (SMILES)-based recurrent neural networks (RNNs) and molecular graph convolutional networks (GCNs) to accurately predict inhibitory activity targeting these 8 kinases.
Collaboration of AI Agents via Cooperative Multi-Agent Deep Reinforcement Learning
Jun 30, 2019



Abstract:There are many AI tasks involving multiple interacting agents where agents should learn to cooperate and collaborate to effectively perform the task. Here we develop and evaluate various multi-agent protocols to train agents to collaborate with teammates in grid soccer. We train and evaluate our multi-agent methods against a team operating with a smart hand-coded policy. As a baseline, we train agents concurrently and independently, with no communication. Our collaborative protocols were parameter sharing, coordinated learning with communication, and counterfactual policy gradients. Against the hand-coded team, the team trained with parameter sharing and the team trained with coordinated learning performed the best, scoring on 89.5% and 94.5% of episodes respectively when playing against the hand-coded team. Against the parameter sharing team, with adversarial training the coordinated learning team scored on 75% of the episodes, indicating it is the most adaptable of our methods. The insights gained from our work can be applied to other domains where multi-agent collaboration could be beneficial.
Institutionally Distributed Deep Learning Networks
Sep 10, 2017



Abstract:Deep learning has become a promising approach for automated medical diagnoses. When medical data samples are limited, collaboration among multiple institutions is necessary to achieve high algorithm performance. However, sharing patient data often has limitations due to technical, legal, or ethical concerns. In such cases, sharing a deep learning model is a more attractive alternative. The best method of performing such a task is unclear, however. In this study, we simulate the dissemination of learning deep learning network models across four institutions using various heuristics and compare the results with a deep learning model trained on centrally hosted patient data. The heuristics investigated include ensembling single institution models, single weight transfer, and cyclical weight transfer. We evaluated these approaches for image classification in three independent image collections (retinal fundus photos, mammography, and ImageNet). We find that cyclical weight transfer resulted in a performance (testing accuracy = 77.3%) that was closest to that of centrally hosted patient data (testing accuracy = 78.7%). We also found that there is an improvement in the performance of cyclical weight transfer heuristic with high frequency of weight transfer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge