Yen Nhi Truong Vu
M&M: Tackling False Positives in Mammography with a Multi-view and Multi-instance Learning Sparse Detector
Aug 11, 2023Abstract:Deep-learning-based object detection methods show promise for improving screening mammography, but high rates of false positives can hinder their effectiveness in clinical practice. To reduce false positives, we identify three challenges: (1) unlike natural images, a malignant mammogram typically contains only one malignant finding; (2) mammography exams contain two views of each breast, and both views ought to be considered to make a correct assessment; (3) most mammograms are negative and do not contain any findings. In this work, we tackle the three aforementioned challenges by: (1) leveraging Sparse R-CNN and showing that sparse detectors are more appropriate than dense detectors for mammography; (2) including a multi-view cross-attention module to synthesize information from different views; (3) incorporating multi-instance learning (MIL) to train with unannotated images and perform breast-level classification. The resulting model, M&M, is a Multi-view and Multi-instance learning system that can both localize malignant findings and provide breast-level predictions. We validate M&M's detection and classification performance using five mammography datasets. In addition, we demonstrate the effectiveness of each proposed component through comprehensive ablation studies.
Problems and shortcuts in deep learning for screening mammography
Mar 29, 2023Abstract:This work reveals undiscovered challenges in the performance and generalizability of deep learning models. We (1) identify spurious shortcuts and evaluation issues that can inflate performance and (2) propose training and analysis methods to address them. We trained an AI model to classify cancer on a retrospective dataset of 120,112 US exams (3,467 cancers) acquired from 2008 to 2017 and 16,693 UK exams (5,655 cancers) acquired from 2011 to 2015. We evaluated on a screening mammography test set of 11,593 US exams (102 cancers; 7,594 women; age 57.1 \pm 11.0) and 1,880 UK exams (590 cancers; 1,745 women; age 63.3 \pm 7.2). A model trained on images of only view markers (no breast) achieved a 0.691 AUC. The original model trained on both datasets achieved a 0.945 AUC on the combined US+UK dataset but paradoxically only 0.838 and 0.892 on the US and UK datasets, respectively. Sampling cancers equally from both datasets during training mitigated this shortcut. A similar AUC paradox (0.903) occurred when evaluating diagnostic exams vs screening exams (0.862 vs 0.861, respectively). Removing diagnostic exams during training alleviated this bias. Finally, the model did not exhibit the AUC paradox over scanner models but still exhibited a bias toward Selenia Dimension (SD) over Hologic Selenia (HS) exams. Analysis showed that this AUC paradox occurred when a dataset attribute had values with a higher cancer prevalence (dataset bias) and the model consequently assigned a higher probability to these attribute values (model bias). Stratification and balancing cancer prevalence can mitigate shortcuts during evaluation. Dataset and model bias can introduce shortcuts and the AUC paradox, potentially pervasive issues within the healthcare AI space. Our methods can verify and mitigate shortcuts while providing a clear understanding of performance.
Deep is a Luxury We Don't Have
Aug 11, 2022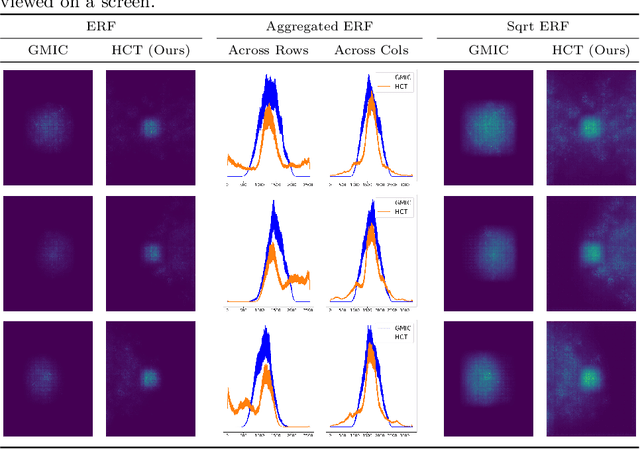
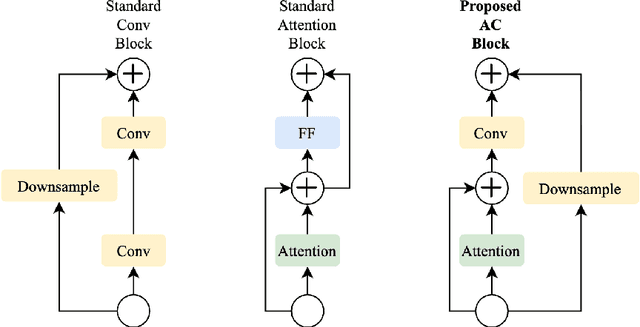
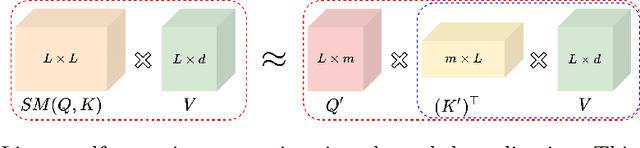

Abstract:Medical images come in high resolutions. A high resolution is vital for finding malignant tissues at an early stage. Yet, this resolution presents a challenge in terms of modeling long range dependencies. Shallow transformers eliminate this problem, but they suffer from quadratic complexity. In this paper, we tackle this complexity by leveraging a linear self-attention approximation. Through this approximation, we propose an efficient vision model called HCT that stands for High resolution Convolutional Transformer. HCT brings transformers' merits to high resolution images at a significantly lower cost. We evaluate HCT using a high resolution mammography dataset. HCT is significantly superior to its CNN counterpart. Furthermore, we demonstrate HCT's fitness for medical images by evaluating its effective receptive field.Code available at https://bit.ly/3ykBhhf
A deep learning algorithm for reducing false positives in screening mammography
Apr 13, 2022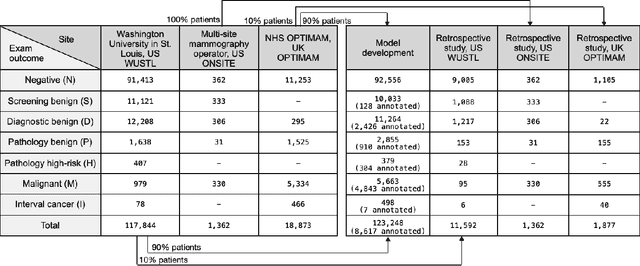
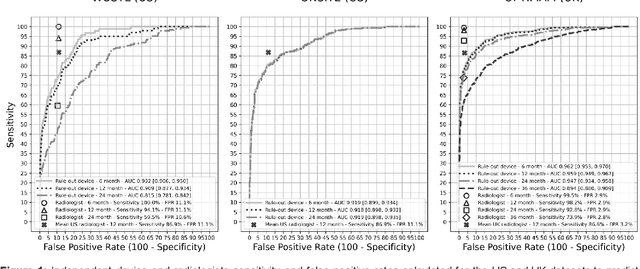
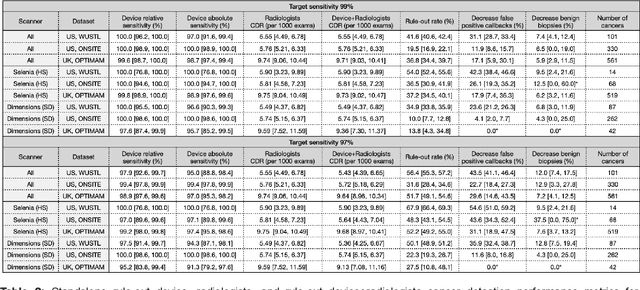
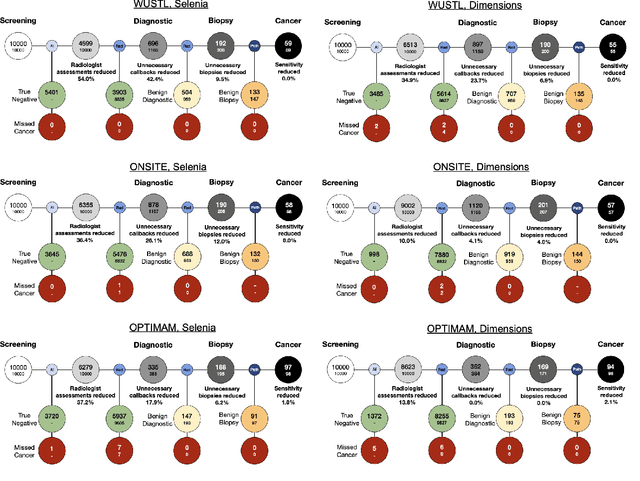
Abstract:Screening mammography improves breast cancer outcomes by enabling early detection and treatment. However, false positive callbacks for additional imaging from screening exams cause unnecessary procedures, patient anxiety, and financial burden. This work demonstrates an AI algorithm that reduces false positives by identifying mammograms not suspicious for breast cancer. We trained the algorithm to determine the absence of cancer using 123,248 2D digital mammograms (6,161 cancers) and performed a retrospective study on 14,831 screening exams (1,026 cancers) from 15 US and 3 UK sites. Retrospective evaluation of the algorithm on the largest of the US sites (11,592 mammograms, 101 cancers) a) left the cancer detection rate unaffected (p=0.02, non-inferiority margin 0.25 cancers per 1000 exams), b) reduced callbacks for diagnostic exams by 31.1% compared to standard clinical readings, c) reduced benign needle biopsies by 7.4%, and d) reduced screening exams requiring radiologist interpretation by 41.6% in the simulated clinical workflow. This work lays the foundation for semi-autonomous breast cancer screening systems that could benefit patients and healthcare systems by reducing false positives, unnecessary procedures, patient anxiety, and expenses.
MedAug: Contrastive learning leveraging patient metadata improves representations for chest X-ray interpretation
Feb 21, 2021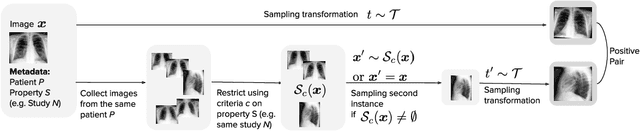
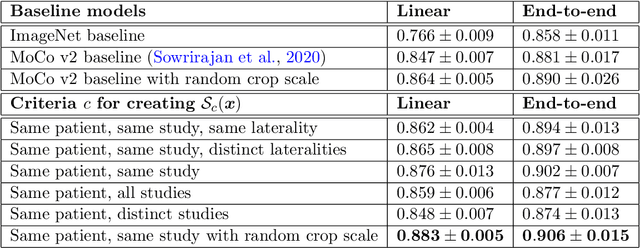

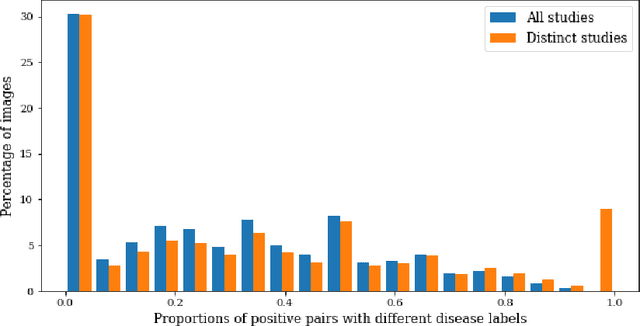
Abstract:Self-supervised contrastive learning between pairs of multiple views of the same image has been shown to successfully leverage unlabeled data to produce meaningful visual representations for both natural and medical images. However, there has been limited work on determining how to select pairs for medical images, where availability of patient metadata can be leveraged to improve representations. In this work, we develop a method to select positive pairs coming from views of possibly different images through the use of patient metadata. We compare strategies for selecting positive pairs for chest X-ray interpretation including requiring them to be from the same patient, imaging study or laterality. We evaluate downstream task performance by fine-tuning the linear layer on 1% of the labeled dataset for pleural effusion classification. Our best performing positive pair selection strategy, which involves using images from the same patient from the same study across all lateralities, achieves a performance increase of 3.4% and 14.4% in mean AUC from both a previous contrastive method and ImageNet pretrained baseline respectively. Our controlled experiments show that the keys to improving downstream performance on disease classification are (1) using patient metadata to appropriately create positive pairs from different images with the same underlying pathologies, and (2) maximizing the number of different images used in query pairing. In addition, we explore leveraging patient metadata to select hard negative pairs for contrastive learning, but do not find improvement over baselines that do not use metadata. Our method is broadly applicable to medical image interpretation and allows flexibility for incorporating medical insights in choosing pairs for contrastive learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge