Darvin Yi
UpBench: A Dynamically Evolving Real-World Labor-Market Agentic Benchmark Framework Built for Human-Centric AI
Nov 15, 2025Abstract:As large language model (LLM) agents increasingly undertake digital work, reliable frameworks are needed to evaluate their real-world competence, adaptability, and capacity for human collaboration. Existing benchmarks remain largely static, synthetic, or domain-limited, providing limited insight into how agents perform in dynamic, economically meaningful environments. We introduce UpBench, a dynamically evolving benchmark grounded in real jobs drawn from the global Upwork labor marketplace. Each task corresponds to a verified client transaction, anchoring evaluation in genuine work activity and financial outcomes. UpBench employs a rubric-based evaluation framework, in which expert freelancers decompose each job into detailed, verifiable acceptance criteria and assess AI submissions with per-criterion feedback. This structure enables fine-grained analysis of model strengths, weaknesses, and instruction-following fidelity beyond binary pass/fail metrics. Human expertise is integrated throughout the data pipeline (from job curation and rubric construction to evaluation) ensuring fidelity to real professional standards and supporting research on human-AI collaboration. By regularly refreshing tasks to reflect the evolving nature of online work, UpBench provides a scalable, human-centered foundation for evaluating agentic systems in authentic labor-market contexts, offering a path toward a collaborative framework, where AI amplifies human capability through partnership rather than replacement.
Targeted Unlearning Using Perturbed Sign Gradient Methods With Applications On Medical Images
May 28, 2025Abstract:Machine unlearning aims to remove the influence of specific training samples from a trained model without full retraining. While prior work has largely focused on privacy-motivated settings, we recast unlearning as a general-purpose tool for post-deployment model revision. Specifically, we focus on utilizing unlearning in clinical contexts where data shifts, device deprecation, and policy changes are common. To this end, we propose a bilevel optimization formulation of boundary-based unlearning that can be solved using iterative algorithms. We provide convergence guarantees when first-order algorithms are used to unlearn. Our method introduces tunable loss design for controlling the forgetting-retention tradeoff and supports novel model composition strategies that merge the strengths of distinct unlearning runs. Across benchmark and real-world clinical imaging datasets, our approach outperforms baselines on both forgetting and retention metrics, including scenarios involving imaging devices and anatomical outliers. This work establishes machine unlearning as a modular, practical alternative to retraining for real-world model maintenance in clinical applications.
Trends, Challenges, and Future Directions in Deep Learning for Glaucoma: A Systematic Review
Nov 07, 2024Abstract:Here, we examine the latest advances in glaucoma detection through Deep Learning (DL) algorithms using Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). This study focuses on three aspects of DL-based glaucoma detection frameworks: input data modalities, processing strategies, and model architectures and applications. Moreover, we analyze trends in employing each aspect since the onset of DL in this field. Finally, we address current challenges and suggest future research directions.
Open-Source Periorbital Segmentation Dataset for Ophthalmic Applications
Sep 30, 2024



Abstract:Periorbital segmentation and distance prediction using deep learning allows for the objective quantification of disease state, treatment monitoring, and remote medicine. However, there are currently no reports of segmentation datasets for the purposes of training deep learning models with sub mm accuracy on the regions around the eyes. All images (n=2842) had the iris, sclera, lid, caruncle, and brow segmented by five trained annotators. Here, we validate this dataset through intra and intergrader reliability tests and show the utility of the data in training periorbital segmentation networks. All the annotations are publicly available for free download. Having access to segmentation datasets designed specifically for oculoplastic surgery will permit more rapid development of clinically useful segmentation networks which can be leveraged for periorbital distance prediction and disease classification. In addition to the annotations, we also provide an open-source toolkit for periorbital distance prediction from segmentation masks. The weights of all models have also been open-sourced and are publicly available for use by the community.
State-of-the-Art Periorbital Distance Prediction and Disease Classification Using Periorbital Features
Sep 27, 2024



Abstract:Periorbital distances and features around the eyes and lids hold valuable information for disease quantification and monitoring of surgical and medical intervention. These distances are commonly measured manually, a process that is both subjective and highly time-consuming. Here, we set out to developed three deep-learning methods for segmentation and periorbital distance prediction, and also evaluate the utility of periorbital distances for disease classification. The MAE of our deep learning predicted distances was less than or very close to the error observed between trained human annotators. We compared our models to the current state-of-the-art (SOTA) method for periorbital distance prediction and found that our methods outperformed SOTA on all of our datasets on all but one periorbital measurement. We also show that robust segmentation can be achieved on diseased eyes using models trained on open-source, healthy eyes, and that periorbital distances have can be used as high-quality features in downstream classification models. Leveraging segmentation networks as intermediary steps in classification has broad implications for increasing the generalizability of classification models in ophthalmic plastic and craniofacial surgery by avoiding the out-of-distribution problem observed in traditional convolutional neural networks.
CvS: Classification via Segmentation For Small Datasets
Oct 29, 2021
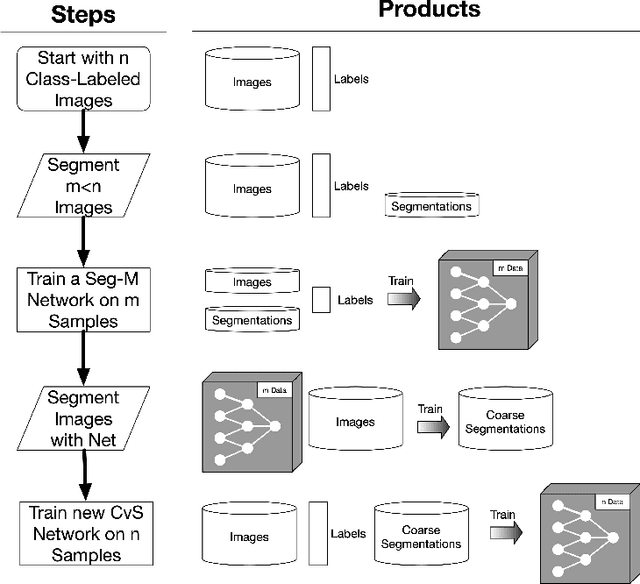
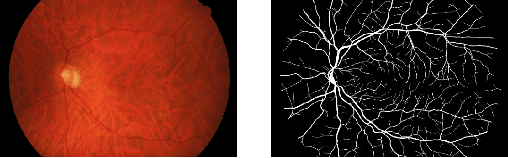
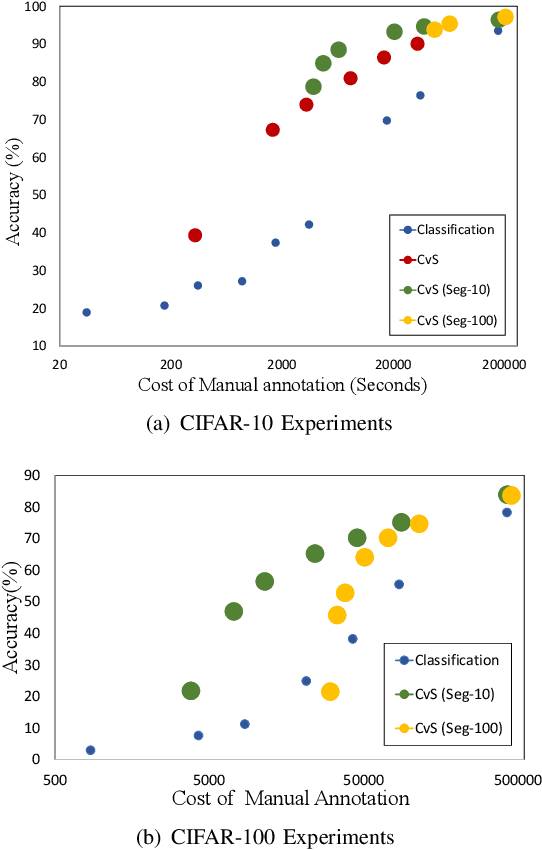
Abstract:Deep learning models have shown promising results in a wide range of computer vision applications across various domains. The success of deep learning methods relies heavily on the availability of a large amount of data. Deep neural networks are prone to overfitting when data is scarce. This problem becomes even more severe for neural network with classification head with access to only a few data points. However, acquiring large-scale datasets is very challenging, laborious, or even infeasible in some domains. Hence, developing classifiers that are able to perform well in small data regimes is crucial for applications with limited data. This paper presents CvS, a cost-effective classifier for small datasets that derives the classification labels from predicting the segmentation maps. We employ the label propagation method to achieve a fully segmented dataset with only a handful of manually segmented data. We evaluate the effectiveness of our framework on diverse problems showing that CvS is able to achieve much higher classification results compared to previous methods when given only a handful of examples.
AutoPtosis
Jun 09, 2021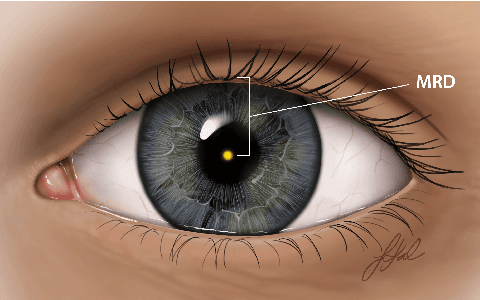

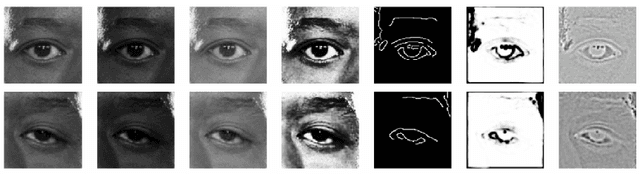

Abstract:Blepharoptosis, or ptosis as it is more commonly referred to, is a condition of the eyelid where the upper eyelid droops. The current diagnosis for ptosis involves cumbersome manual measurements that are time-consuming and prone to human error. In this paper, we present AutoPtosis, an artificial intelligence based system with interpretable results for rapid diagnosis of ptosis. We utilize a diverse dataset collected from the Illinois Ophthalmic Database Atlas (I-ODA) to develop a robust deep learning model for prediction and also develop a clinically inspired model that calculates the marginal reflex distance and iris ratio. AutoPtosis achieved 95.5% accuracy on physician verified data that had an equal class balance. The proposed algorithm can help in the rapid and timely diagnosis of ptosis, significantly reduce the burden on the healthcare system, and save the patients and clinics valuable resources.
I-ODA, Real-World Multi-modal Longitudinal Data for OphthalmicApplications
Mar 30, 2021
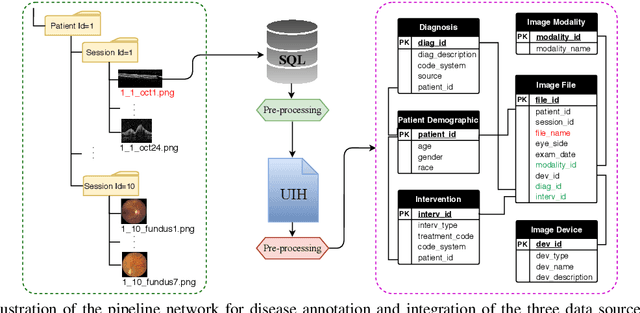
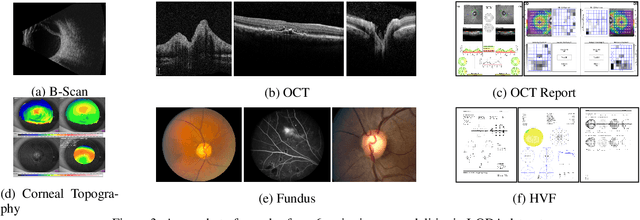
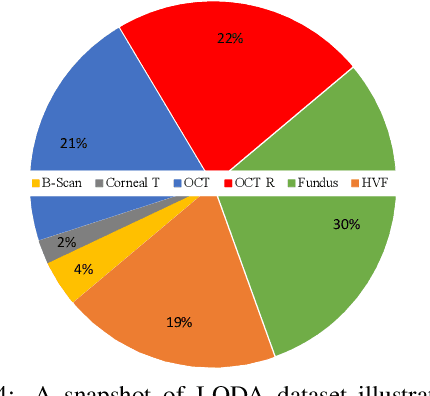
Abstract:Data from clinical real-world settings is characterized by variability in quality, machine-type, setting, and source. One of the primary goals of medical computer vision is to develop and validate artificial intelligence (AI) based algorithms on real-world data enabling clinical translations. However, despite the exponential growth in AI based applications in healthcare, specifically in ophthalmology, translations to clinical settings remain challenging. Limited access to adequate and diverse real-world data inhibits the development and validation of translatable algorithms. In this paper, we present a new multi-modal longitudinal ophthalmic imaging dataset, the Illinois Ophthalmic Database Atlas (I-ODA), with the goal of advancing state-of-the-art computer vision applications in ophthalmology, and improving upon the translatable capacity of AI based applications across different clinical settings. We present the infrastructure employed to collect, annotate, and anonymize images from multiple sources, demonstrating the complexity of real-world retrospective data and its limitations. I-ODA includes 12 imaging modalities with a total of 3,668,649 ophthalmic images of 33,876 individuals from the Department of Ophthalmology and Visual Sciences at the Illinois Eye and Ear Infirmary of the University of Illinois Chicago (UIC) over the course of 12 years.
Real-World Multi-Domain Data Applications for Generalizations to Clinical Settings
Jul 24, 2020
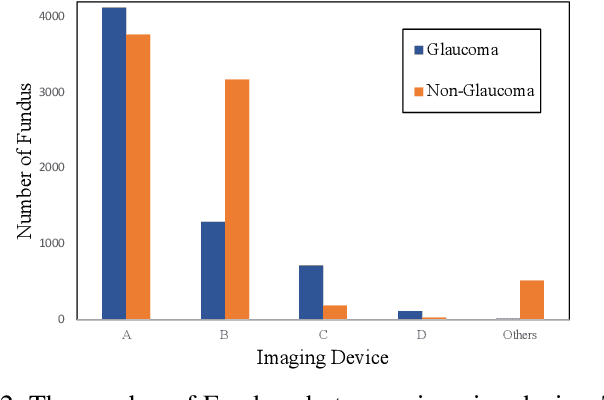
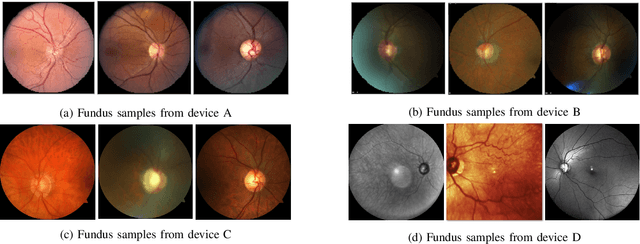

Abstract:With promising results of machine learning based models in computer vision, applications on medical imaging data have been increasing exponentially. However, generalizations to complex real-world clinical data is a persistent problem. Deep learning models perform well when trained on standardized datasets from artificial settings, such as clinical trials. However, real-world data is different and translations are yielding varying results. The complexity of real-world applications in healthcare could emanate from a mixture of different data distributions across multiple device domains alongside the inevitable noise sourced from varying image resolutions, human errors, and the lack of manual gradings. In addition, healthcare applications not only suffer from the scarcity of labeled data, but also face limited access to unlabeled data due to HIPAA regulations, patient privacy, ambiguity in data ownership, and challenges in collecting data from different sources. These limitations pose additional challenges to applying deep learning algorithms in healthcare and clinical translations. In this paper, we utilize self-supervised representation learning methods, formulated effectively in transfer learning settings, to address limited data availability. Our experiments verify the importance of diverse real-world data for generalization to clinical settings. We show that by employing a self-supervised approach with transfer learning on a multi-domain real-world dataset, we can achieve 16% relative improvement on a standardized dataset over supervised baselines.
Random Bundle: Brain Metastases Segmentation Ensembling through Annotation Randomization
Feb 23, 2020



Abstract:We introduce a novel ensembling method, Random Bundle (RB), that improves performance for brain metastases segmentation. We create our ensemble by training each network on our dataset with 50% of our annotated lesions censored out. We also apply a lopsided bootstrap loss to recover performance after inducing an in silico 50% false negative rate and make our networks more sensitive. We improve our network detection of lesions's mAP value by 39% and more than triple the sensitivity at 80% precision. We also show slight improvements in segmentation quality through DICE score. Further, RB ensembling improves performance over baseline by a larger margin than a variety of popular ensembling strategies. Finally, we show that RB ensembling is computationally efficient by comparing its performance to a single network when both systems are constrained to have the same compute.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge