Abdullah Aleem
AutoPtosis
Jun 09, 2021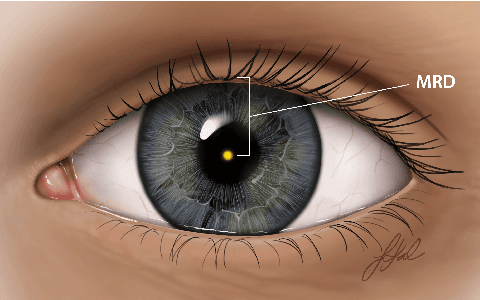


Abstract:Blepharoptosis, or ptosis as it is more commonly referred to, is a condition of the eyelid where the upper eyelid droops. The current diagnosis for ptosis involves cumbersome manual measurements that are time-consuming and prone to human error. In this paper, we present AutoPtosis, an artificial intelligence based system with interpretable results for rapid diagnosis of ptosis. We utilize a diverse dataset collected from the Illinois Ophthalmic Database Atlas (I-ODA) to develop a robust deep learning model for prediction and also develop a clinically inspired model that calculates the marginal reflex distance and iris ratio. AutoPtosis achieved 95.5% accuracy on physician verified data that had an equal class balance. The proposed algorithm can help in the rapid and timely diagnosis of ptosis, significantly reduce the burden on the healthcare system, and save the patients and clinics valuable resources.
I-ODA, Real-World Multi-modal Longitudinal Data for OphthalmicApplications
Mar 30, 2021
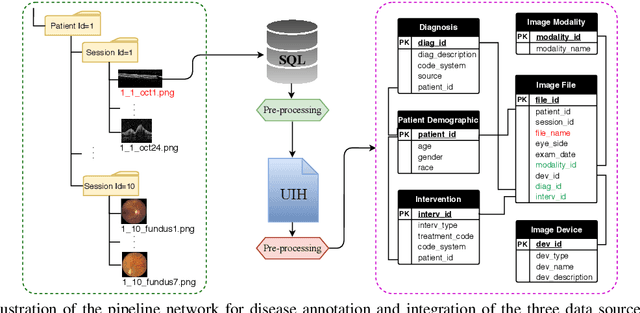
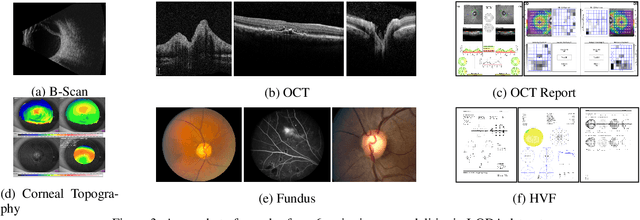
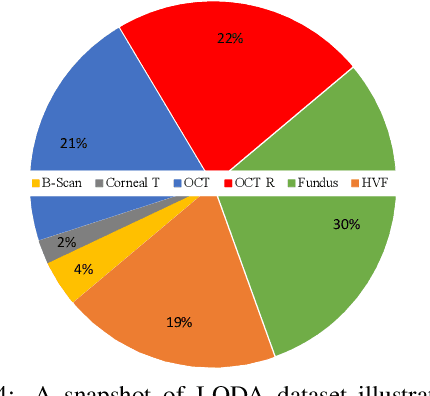
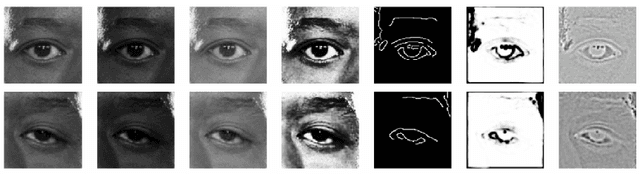
Abstract:Data from clinical real-world settings is characterized by variability in quality, machine-type, setting, and source. One of the primary goals of medical computer vision is to develop and validate artificial intelligence (AI) based algorithms on real-world data enabling clinical translations. However, despite the exponential growth in AI based applications in healthcare, specifically in ophthalmology, translations to clinical settings remain challenging. Limited access to adequate and diverse real-world data inhibits the development and validation of translatable algorithms. In this paper, we present a new multi-modal longitudinal ophthalmic imaging dataset, the Illinois Ophthalmic Database Atlas (I-ODA), with the goal of advancing state-of-the-art computer vision applications in ophthalmology, and improving upon the translatable capacity of AI based applications across different clinical settings. We present the infrastructure employed to collect, annotate, and anonymize images from multiple sources, demonstrating the complexity of real-world retrospective data and its limitations. I-ODA includes 12 imaging modalities with a total of 3,668,649 ophthalmic images of 33,876 individuals from the Department of Ophthalmology and Visual Sciences at the Illinois Eye and Ear Infirmary of the University of Illinois Chicago (UIC) over the course of 12 years.
Real-World Multi-Domain Data Applications for Generalizations to Clinical Settings
Jul 24, 2020
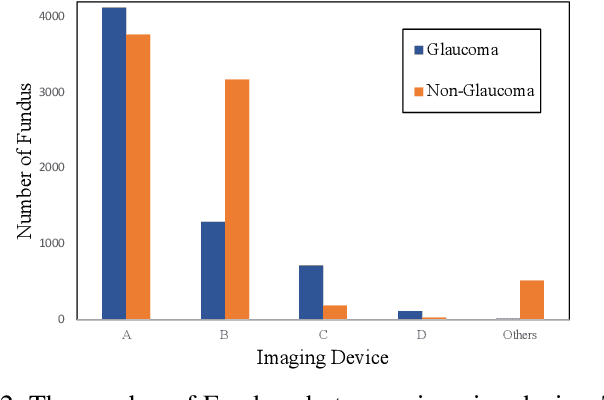
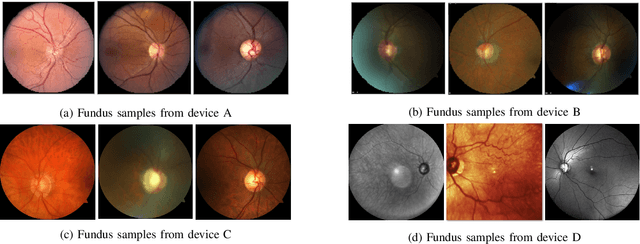

Abstract:With promising results of machine learning based models in computer vision, applications on medical imaging data have been increasing exponentially. However, generalizations to complex real-world clinical data is a persistent problem. Deep learning models perform well when trained on standardized datasets from artificial settings, such as clinical trials. However, real-world data is different and translations are yielding varying results. The complexity of real-world applications in healthcare could emanate from a mixture of different data distributions across multiple device domains alongside the inevitable noise sourced from varying image resolutions, human errors, and the lack of manual gradings. In addition, healthcare applications not only suffer from the scarcity of labeled data, but also face limited access to unlabeled data due to HIPAA regulations, patient privacy, ambiguity in data ownership, and challenges in collecting data from different sources. These limitations pose additional challenges to applying deep learning algorithms in healthcare and clinical translations. In this paper, we utilize self-supervised representation learning methods, formulated effectively in transfer learning settings, to address limited data availability. Our experiments verify the importance of diverse real-world data for generalization to clinical settings. We show that by employing a self-supervised approach with transfer learning on a multi-domain real-world dataset, we can achieve 16% relative improvement on a standardized dataset over supervised baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge