Elizabeth Tong
Beyond the Hype: Assessing the Performance, Trustworthiness, and Clinical Suitability of GPT3.5
Jun 28, 2023
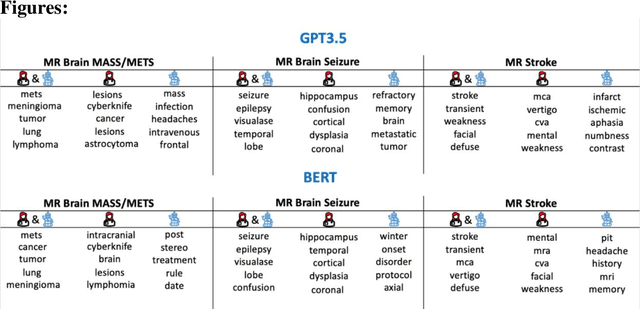
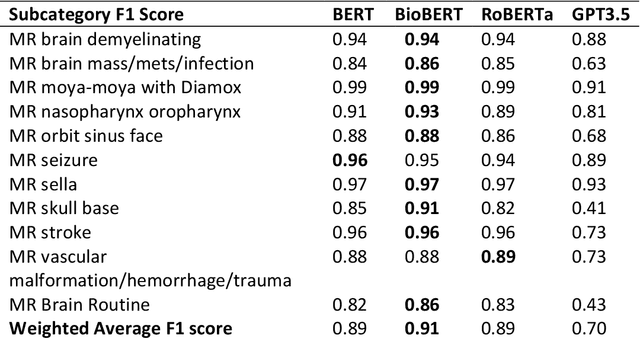
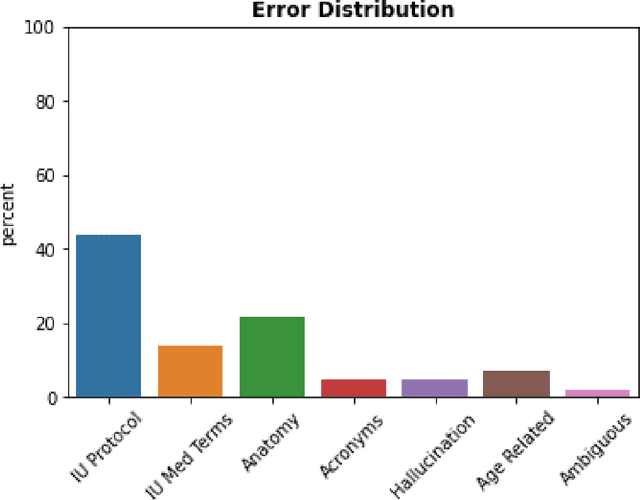
Abstract:The use of large language models (LLMs) in healthcare is gaining popularity, but their practicality and safety in clinical settings have not been thoroughly assessed. In high-stakes environments like medical settings, trust and safety are critical issues for LLMs. To address these concerns, we present an approach to evaluate the performance and trustworthiness of a GPT3.5 model for medical image protocol assignment. We compare it with a fine-tuned BERT model and a radiologist. In addition, we have a radiologist review the GPT3.5 output to evaluate its decision-making process. Our evaluation dataset consists of 4,700 physician entries across 11 imaging protocol classes spanning the entire head. Our findings suggest that the GPT3.5 performance falls behind BERT and a radiologist. However, GPT3.5 outperforms BERT in its ability to explain its decision, detect relevant word indicators, and model calibration. Furthermore, by analyzing the explanations of GPT3.5 for misclassifications, we reveal systematic errors that need to be resolved to enhance its safety and suitability for clinical use.
Random Bundle: Brain Metastases Segmentation Ensembling through Annotation Randomization
Feb 23, 2020



Abstract:We introduce a novel ensembling method, Random Bundle (RB), that improves performance for brain metastases segmentation. We create our ensemble by training each network on our dataset with 50% of our annotated lesions censored out. We also apply a lopsided bootstrap loss to recover performance after inducing an in silico 50% false negative rate and make our networks more sensitive. We improve our network detection of lesions's mAP value by 39% and more than triple the sensitivity at 80% precision. We also show slight improvements in segmentation quality through DICE score. Further, RB ensembling improves performance over baseline by a larger margin than a variety of popular ensembling strategies. Finally, we show that RB ensembling is computationally efficient by comparing its performance to a single network when both systems are constrained to have the same compute.
Brain Metastasis Segmentation Network Trained with Robustness to Annotations with Multiple False Negatives
Jan 26, 2020



Abstract:Deep learning has proven to be an essential tool for medical image analysis. However, the need for accurately labeled input data, often requiring time- and labor-intensive annotation by experts, is a major limitation to the use of deep learning. One solution to this challenge is to allow for use of coarse or noisy labels, which could permit more efficient and scalable labeling of images. In this work, we develop a lopsided loss function based on entropy regularization that assumes the existence of a nontrivial false negative rate in the target annotations. Starting with a carefully annotated brain metastasis lesion dataset, we simulate data with false negatives by (1) randomly censoring the annotated lesions and (2) systematically censoring the smallest lesions. The latter better models true physician error because smaller lesions are harder to notice than the larger ones. Even with a simulated false negative rate as high as 50%, applying our loss function to randomly censored data preserves maximum sensitivity at 97% of the baseline with uncensored training data, compared to just 10% for a standard loss function. For the size-based censorship, performance is restored from 17% with the current standard to 88% with our lopsided bootstrap loss. Our work will enable more efficient scaling of the image labeling process, in parallel with other approaches on creating more efficient user interfaces and tools for annotation.
Handling Missing MRI Input Data in Deep Learning Segmentation of Brain Metastases: A Multi-Center Study
Dec 27, 2019



Abstract:The purpose was to assess the clinical value of a novel DropOut model for detecting and segmenting brain metastases, in which a neural network is trained on four distinct MRI sequences using an input dropout layer, thus simulating the scenario of missing MRI data by training on the full set and all possible subsets of the input data. This retrospective, multi-center study, evaluated 165 patients with brain metastases. A deep learning based segmentation model for automatic segmentation of brain metastases, named DropOut, was trained on multi-sequence MRI from 100 patients, and validated/tested on 10/55 patients. The segmentation results were compared with the performance of a state-of-the-art DeepLabV3 model. The MR sequences in the training set included pre- and post-gadolinium (Gd) T1-weighted 3D fast spin echo, post-Gd T1-weighted inversion recovery (IR) prepped fast spoiled gradient echo, and 3D fluid attenuated inversion recovery (FLAIR), whereas the test set did not include the IR prepped image-series. The ground truth were established by experienced neuroradiologists. The results were evaluated using precision, recall, Dice score, and receiver operating characteristics (ROC) curve statistics, while the Wilcoxon rank sum test was used to compare the performance of the two neural networks. The area under the ROC curve (AUC), averaged across all test cases, was 0.989+-0.029 for the DropOut model and 0.989+-0.023 for the DeepLabV3 model (p=0.62). The DropOut model showed a significantly higher Dice score compared to the DeepLabV3 model (0.795+-0.105 vs. 0.774+-0.104, p=0.017), and a significantly lower average false positive rate of 3.6/patient vs. 7.0/patient (p<0.001) using a 10mm3 lesion-size limit. The DropOut model may facilitate accurate detection and segmentation of brain metastases on a multi-center basis, even when the test cohort is missing MRI input data.
MRI Pulse Sequence Integration for Deep-Learning Based Brain Metastasis Segmentation
Dec 18, 2019



Abstract:Magnetic resonance (MR) imaging is an essential diagnostic tool in clinical medicine. Recently, a variety of deep learning methods have been applied to segmentation tasks in medical images, with promising results for computer-aided diagnosis. For MR images, effectively integrating different pulse sequences is important to optimize performance. However, the best way to integrate different pulse sequences remains unclear. In this study, we evaluate multiple architectural features and characterize their effects in the task of metastasis segmentation. Specifically, we consider (1) different pulse sequence integration schemas, (2) different modes of weight sharing for parallel network branches, and (3) a new approach for enabling robustness to missing pulse sequences. We find that levels of integration and modes of weight sharing that favor low variance work best in our regime of small data (n = 100). By adding an input-level dropout layer, we could preserve the overall performance of these networks while allowing for inference on inputs with missing pulse sequence. We illustrate not only the generalizability of the network but also the utility of this robustness when applying the trained model to data from a different center, which does not use the same pulse sequences. Finally, we apply network visualization methods to better understand which input features are most important for network performance. Together, these results provide a framework for building networks with enhanced robustness to missing data while maintaining comparable performance in medical imaging applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge