Prediction of Small Molecule Kinase Inhibitors for Chemotherapy Using Deep Learning
Paper and Code
Jun 30, 2019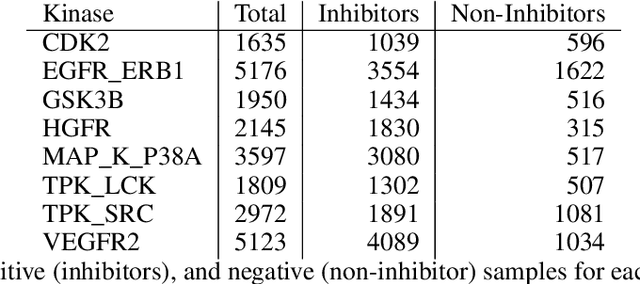
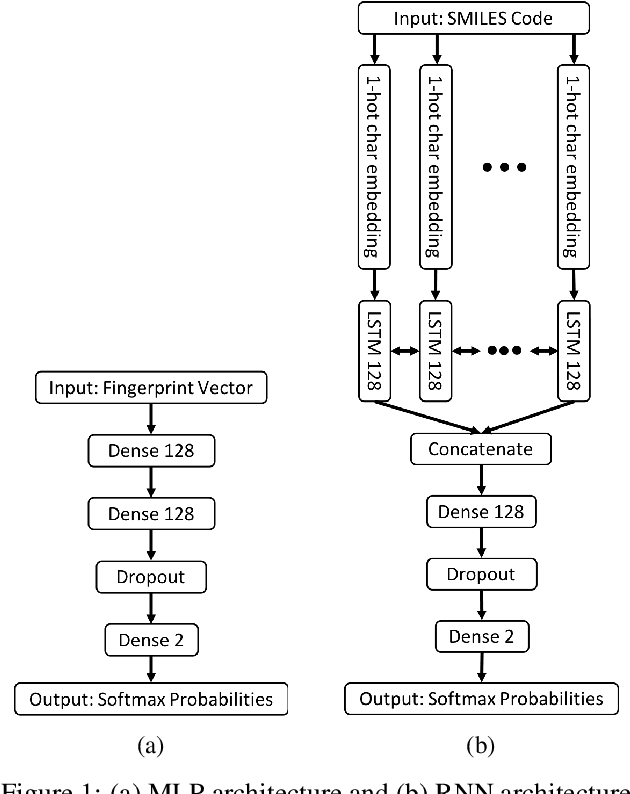
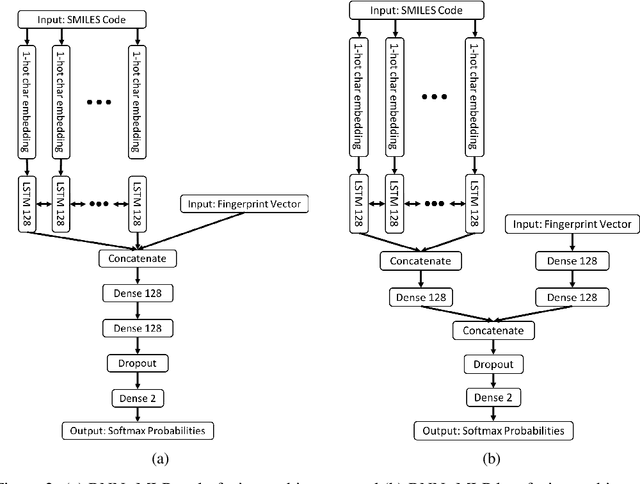
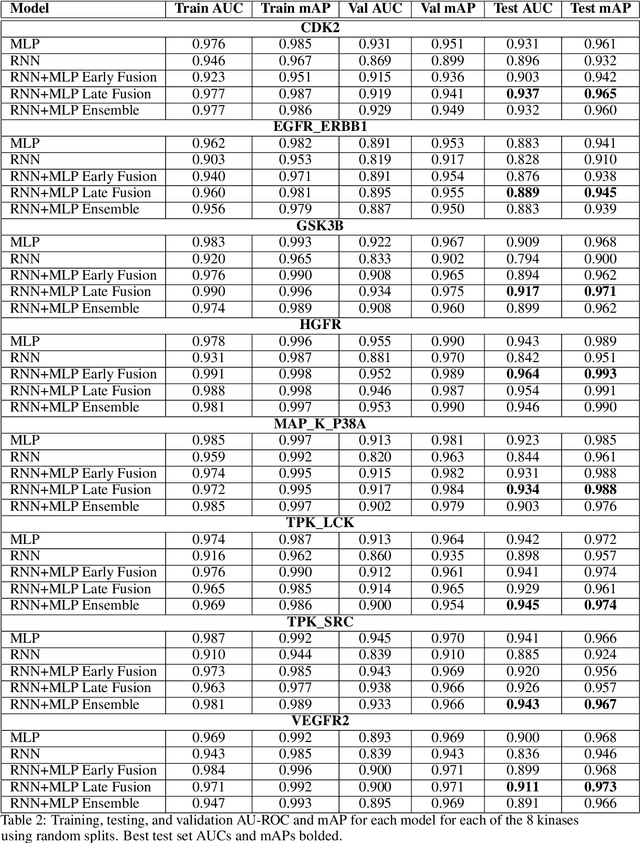
The current state of cancer therapeutics has been moving away from one-size-fits-all cytotoxic chemotherapy, and towards a more individualized and specific approach involving the targeting of each tumor's genetic vulnerabilities. Different tumors, even of the same type, may be more reliant on certain cellular pathways more than others. With modern advancements in our understanding of cancer genome sequencing, these pathways can be discovered. Investigating each of the millions of possible small molecule inhibitors for each kinase in vitro, however, would be extremely expensive and time consuming. This project focuses on predicting the inhibition activity of small molecules targeting 8 different kinases using multiple deep learning models. We trained fingerprint-based MLPs and simplified molecular-input line-entry specification (SMILES)-based recurrent neural networks (RNNs) and molecular graph convolutional networks (GCNs) to accurately predict inhibitory activity targeting these 8 kinases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge