Niloy Ganguly
IDALC: A Semi-Supervised Framework for Intent Detection and Active Learning based Correction
Nov 08, 2025



Abstract:Voice-controlled dialog systems have become immensely popular due to their ability to perform a wide range of actions in response to diverse user queries. These agents possess a predefined set of skills or intents to fulfill specific user tasks. But every system has its own limitations. There are instances where, even for known intents, if any model exhibits low confidence, it results in rejection of utterances that necessitate manual annotation. Additionally, as time progresses, there may be a need to retrain these agents with new intents from the system-rejected queries to carry out additional tasks. Labeling all these emerging intents and rejected utterances over time is impractical, thus calling for an efficient mechanism to reduce annotation costs. In this paper, we introduce IDALC (Intent Detection and Active Learning based Correction), a semi-supervised framework designed to detect user intents and rectify system-rejected utterances while minimizing the need for human annotation. Empirical findings on various benchmark datasets demonstrate that our system surpasses baseline methods, achieving a 5-10% higher accuracy and a 4-8% improvement in macro-F1. Remarkably, we maintain the overall annotation cost at just 6-10% of the unlabelled data available to the system. The overall framework of IDALC is shown in Fig. 1
* Paper accepted in IEEE Transactions on Artificial Intelligence (October 2025)
LLM Meets Diffusion: A Hybrid Framework for Crystal Material Generation
Oct 27, 2025Abstract:Recent advances in generative modeling have shown significant promise in designing novel periodic crystal structures. Existing approaches typically rely on either large language models (LLMs) or equivariant denoising models, each with complementary strengths: LLMs excel at handling discrete atomic types but often struggle with continuous features such as atomic positions and lattice parameters, while denoising models are effective at modeling continuous variables but encounter difficulties in generating accurate atomic compositions. To bridge this gap, we propose CrysLLMGen, a hybrid framework that integrates an LLM with a diffusion model to leverage their complementary strengths for crystal material generation. During sampling, CrysLLMGen first employs a fine-tuned LLM to produce an intermediate representation of atom types, atomic coordinates, and lattice structure. While retaining the predicted atom types, it passes the atomic coordinates and lattice structure to a pre-trained equivariant diffusion model for refinement. Our framework outperforms state-of-the-art generative models across several benchmark tasks and datasets. Specifically, CrysLLMGen not only achieves a balanced performance in terms of structural and compositional validity but also generates more stable and novel materials compared to LLM-based and denoisingbased models Furthermore, CrysLLMGen exhibits strong conditional generation capabilities, effectively producing materials that satisfy user-defined constraints. Code is available at https://github.com/kdmsit/crysllmgen
Brevity is the soul of sustainability: Characterizing LLM response lengths
Jun 10, 2025



Abstract:A significant portion of the energy consumed by Large Language Models (LLMs) arises from their inference processes; hence developing energy-efficient methods for inference is crucial. While several techniques exist for inference optimization, output compression remains relatively unexplored, with only a few preliminary efforts addressing this aspect. In this work, we first benchmark 12 decoder-only LLMs across 5 datasets, revealing that these models often produce responses that are substantially longer than necessary. We then conduct a comprehensive quality assessment of LLM responses, formally defining six information categories present in LLM responses. We show that LLMs often tend to include redundant or additional information besides the minimal answer. To address this issue of long responses by LLMs, we explore several simple and intuitive prompt-engineering strategies. Empirical evaluation shows that appropriate prompts targeting length reduction and controlling information content can achieve significant energy optimization between 25-60\% by reducing the response length while preserving the quality of LLM responses.
Label-semantics Aware Generative Approach for Domain-Agnostic Multilabel Classification
Jun 07, 2025



Abstract:The explosion of textual data has made manual document classification increasingly challenging. To address this, we introduce a robust, efficient domain-agnostic generative model framework for multi-label text classification. Instead of treating labels as mere atomic symbols, our approach utilizes predefined label descriptions and is trained to generate these descriptions based on the input text. During inference, the generated descriptions are matched to the pre-defined labels using a finetuned sentence transformer. We integrate this with a dual-objective loss function, combining cross-entropy loss and cosine similarity of the generated sentences with the predefined target descriptions, ensuring both semantic alignment and accuracy. Our proposed model LAGAMC stands out for its parameter efficiency and versatility across diverse datasets, making it well-suited for practical applications. We demonstrate the effectiveness of our proposed model by achieving new state-of-the-art performances across all evaluated datasets, surpassing several strong baselines. We achieve improvements of 13.94% in Micro-F1 and 24.85% in Macro-F1 compared to the closest baseline across all datasets.
Evaluation of LLMs in Medical Text Summarization: The Role of Vocabulary Adaptation in High OOV Settings
May 27, 2025



Abstract:Large Language Models (LLMs) recently achieved great success in medical text summarization by simply using in-context learning. However, these recent efforts do not perform fine-grained evaluations under difficult settings where LLMs might fail. They typically report performance scores over the entire dataset. Through our benchmarking study, we show that LLMs show a significant performance drop for data points with high concentration of out-of-vocabulary (OOV) words or with high novelty. Vocabulary adaptation is an intuitive solution to this vocabulary mismatch issue where the LLM vocabulary gets updated with certain expert domain (here, medical) words or subwords. An interesting finding from our study is that Llama-3.1, even with a vocabulary size of around 128K tokens, still faces over-fragmentation issue with medical words. To that end, we show vocabulary adaptation helps improve the LLM summarization performance even in difficult settings. Through extensive experimentation of multiple vocabulary adaptation strategies, two continual pretraining strategies, and three benchmark medical summarization datasets, we gain valuable insights into the role of vocabulary adaptation strategies for customizing LLMs to the medical domain. We also performed a human evaluation study with medical experts where they found that vocabulary adaptation results in more relevant and faithful summaries. Our codebase is made publicly available at https://github.com/gb-kgp/LLM-MedicalSummarization-Benchmark.
Towards Sustainable NLP: Insights from Benchmarking Inference Energy in Large Language Models
Feb 08, 2025



Abstract:Large language models (LLMs) are increasingly recognized for their exceptional generative capabilities and versatility across various tasks. However, the high inference costs associated with these models have not received adequate attention, particularly when compared to the focus on training costs in existing research. In response to this gap, our study conducts a comprehensive benchmarking of LLM inference energy across a wide range of NLP tasks, where we analyze the impact of different models, tasks, prompts, and system-related factors on inference energy. Specifically, our experiments reveal several interesting insights, including strong correlation of inference energy with output token length and response time. Also, we find that quantization and optimal batch sizes, along with targeted prompt phrases, can significantly reduce energy usage. This study is the first to thoroughly benchmark LLM inference across such a diverse range of aspects, providing insights and offering several recommendations for improving energy efficiency in model deployment.
Efficient Continual Pre-training of LLMs for Low-resource Languages
Dec 13, 2024Abstract:Open-source Large Language models (OsLLMs) propel the democratization of natural language research by giving the flexibility to augment or update model parameters for performance improvement. Nevertheless, like proprietary LLMs, Os-LLMs offer poorer performance on low-resource languages (LRLs) than high-resource languages (HRLs), owing to smaller amounts of training data and underrepresented vocabulary. On the other hand, continual pre-training (CPT) with large amounts of language-specific data is a costly proposition in terms of data acquisition and computational resources. Our goal is to drastically reduce CPT cost. To that end, we first develop a new algorithm to select a subset of texts from a larger corpus. We show the effectiveness of our technique using very little CPT data. In search of further improvement, we design a new algorithm to select tokens to include in the LLM vocabulary. We experiment with the recent Llama-3 model and nine Indian languages with diverse scripts and extent of resource availability. For evaluation, we use IndicGenBench, a generation task benchmark dataset for Indic languages. We experiment with various CPT corpora and augmented vocabulary size and offer insights across language families.
Adaptive BPE Tokenization for Enhanced Vocabulary Adaptation in Finetuning Pretrained Language Models
Oct 04, 2024



Abstract:In this work, we show a fundamental limitation in vocabulary adaptation approaches that use Byte-Pair Encoding (BPE) tokenization scheme for fine-tuning pretrained language models (PLMs) to expert domains. Current approaches trivially append the target domain-specific vocabulary at the end of the PLM vocabulary. This approach leads to a lower priority score and causes sub-optimal tokenization in BPE that iteratively uses merge rules to tokenize a given text. To mitigate this issue, we propose AdaptBPE where the BPE tokenization initialization phase is modified to first perform the longest string matching on the added (target) vocabulary before tokenizing at the character level. We perform an extensive evaluation of AdaptBPE versus the standard BPE over various classification and summarization tasks; AdaptBPE improves by 3.57% (in terms of accuracy) and 1.87% (in terms of Rouge-L), respectively. AdaptBPE for MEDVOC works particularly well when reference summaries have high OOV concentration or are longer in length. We also conduct a human evaluation, revealing that AdaptBPE generates more relevant and more faithful summaries as compared to MEDVOC. We make our codebase publicly available at https://github.com/gb-kgp/adaptbpe.
Unlocking Efficiency: Adaptive Masking for Gene Transformer Models
Aug 13, 2024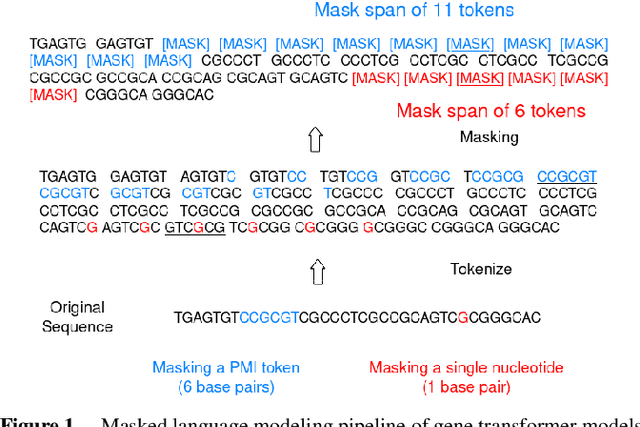
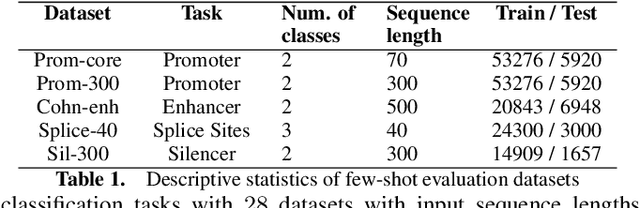
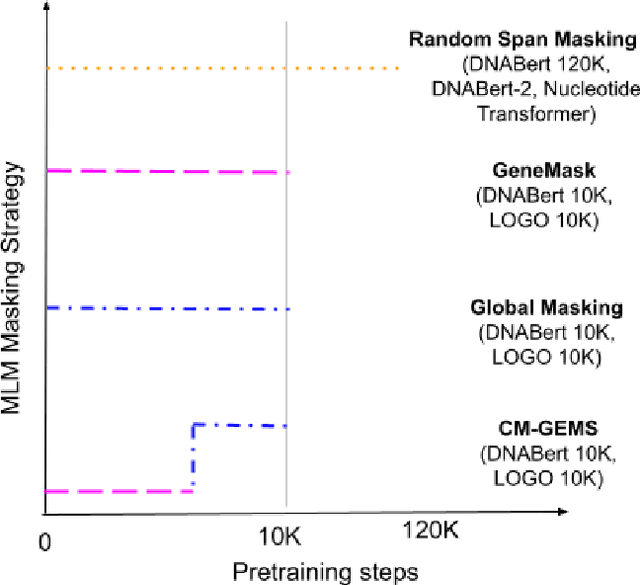
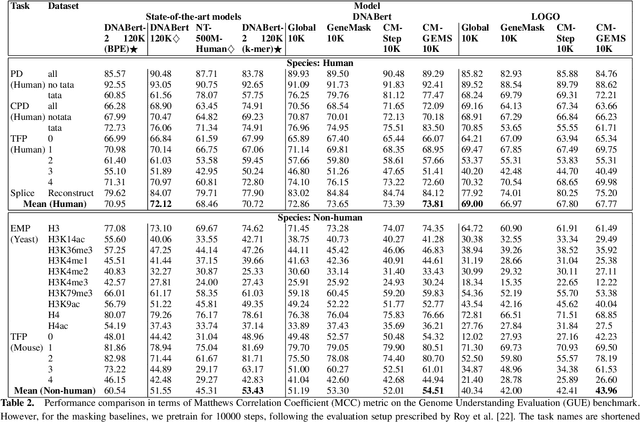
Abstract:Gene transformer models such as Nucleotide Transformer, DNABert, and LOGO are trained to learn optimal gene sequence representations by using the Masked Language Modeling (MLM) training objective over the complete Human Reference Genome. However, the typical tokenization methods employ a basic sliding window of tokens, such as k-mers, that fail to utilize gene-centric semantics. This could result in the (trivial) masking of easily predictable sequences, leading to inefficient MLM training. Time-variant training strategies are known to improve pretraining efficiency in both language and vision tasks. In this work, we focus on using curriculum masking where we systematically increase the difficulty of masked token prediction task by using a Pointwise Mutual Information-based difficulty criterion, as gene sequences lack well-defined semantic units similar to words or sentences of NLP domain. Our proposed Curriculum Masking-based Gene Masking Strategy (CM-GEMS) demonstrates superior representation learning capabilities compared to baseline masking approaches when evaluated on downstream gene sequence classification tasks. We perform extensive evaluation in both few-shot (five datasets) and full dataset settings (Genomic Understanding Evaluation benchmark consisting of 27 tasks). Our findings reveal that CM-GEMS outperforms state-of-the-art models (DNABert-2, Nucleotide transformer, DNABert) trained at 120K steps, achieving similar results in just 10K and 1K steps. We also demonstrate that Curriculum-Learned LOGO (a 2-layer DNABert-like model) can achieve nearly 90% of the state-of-the-art model performance of 120K steps. We will make the models and codes publicly available at https://github.com/roysoumya/curriculum-GeneMask.
Leveraging the Power of LLMs: A Fine-Tuning Approach for High-Quality Aspect-Based Summarization
Aug 05, 2024



Abstract:The ever-increasing volume of digital information necessitates efficient methods for users to extract key insights from lengthy documents. Aspect-based summarization offers a targeted approach, generating summaries focused on specific aspects within a document. Despite advancements in aspect-based summarization research, there is a continuous quest for improved model performance. Given that large language models (LLMs) have demonstrated the potential to revolutionize diverse tasks within natural language processing, particularly in the problem of summarization, this paper explores the potential of fine-tuning LLMs for the aspect-based summarization task. We evaluate the impact of fine-tuning open-source foundation LLMs, including Llama2, Mistral, Gemma and Aya, on a publicly available domain-specific aspect based summary dataset. We hypothesize that this approach will enable these models to effectively identify and extract aspect-related information, leading to superior quality aspect-based summaries compared to the state-of-the-art. We establish a comprehensive evaluation framework to compare the performance of fine-tuned LLMs against competing aspect-based summarization methods and vanilla counterparts of the fine-tuned LLMs. Our work contributes to the field of aspect-based summarization by demonstrating the efficacy of fine-tuning LLMs for generating high-quality aspect-based summaries. Furthermore, it opens doors for further exploration of using LLMs for targeted information extraction tasks across various NLP domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge